Wiki
Clone wikigmc-synthesis-v0.2 / Home
Welcome
This is Emilio's branch of Julien's gmc-synthesis, a tool implementing the theory introduced in From Communicating Machines to Graphical Choreographies (J. Lange, E. Tuosto, and N. Yoshida, POPL 2015). This branch is also based on An abstract semantics of the global view of choreographies (R. Guanciale and E. Tuosto, ICE 2016).
Forewords
This branch has been started in order to
- improve the feedback to the user when generalised multiparty compatibility is violated,
- add new features to
- manipuate the TS (eg highlighting of configurations with some properties; click here for examples of graphical outputs.)
- improve usability
- use asserted CFSMs
- track paths from global graphs to the corresponding TS
(all this is still work in progress).
The main commands provided by the implementation are
- gmc: takes in input a CFSM system, checks it for generalised multiparty compatibility, and builds the corresponding global graph
- chosyn.py: a python script that executes gmc, transforms the .dot files it generates in graphical formats, and displays some performance information
- sgg: takes in input a description of a global graph in the syntax of the ICE 2016 paper and projects the graph in a set of (non-minimal) CFSM provided that the graph is well-branched and well-sequentiased.
Syntax of the input files
Strings are made of the following characters
0123456789<=>ABCDEFGHIJKLMNOPQRSTUVWXYZ()\^_`abcdefghijklmnopqrstuvwxyz\167/$#&~,
and must start with a letter when specifying the name of a machine (non-terminal Ptp).
GMC
The syntax of the input files of gmc can be either in the 'fsa' (after finite state automata) or 'sys' format, the latter being the simple process-algebraic syntax described in the following. Which format is used depends on the extesion to the file name ('.fsa' or '.sys' respectively).
The 'fsa' format is a text where each line describes (some element of) a CFSM: for instance,
#!ABNF .outputs A .state graph q0 1 ! hello q1 q0 1 ! world q1 .marking q0 .end .outputs .state graph q0 0 ? hello q1 q0 0 ? world q1 .marking q0 .end
specifies a system of two machines. - machine 0 is named A (line '.outputs A'), has q0 as initial state (line .marking q0) from which it may send either a message 'hello' or a message 'world' to machine 1 and move to state q1 (lines 'q0 1 ! hello q1' and 'q0 1 ! world q1', respectively). Messages can be any sort for string; the current implementation does not interpret them, but in the future we plan to fix a syntax and a semantics in order to deal with e.g., variables, binderds, values, etc. - machine 1 is ready to receive either message sent by machine 0.
Machine names are optional (e.g., machine 1 is anonymous in the example above); if present in the input file, they are used to denote the channels in the files generated by the tool (for instance, the first transition of A is displayed as "AB ! hello" rather than "01!hello". Lines '.outputs ...', '.state graph', '.marking ...', and '.end' are mandatory. Lines starting with '--' are ignored.
The 'sys' format is probably more appropriate for systems consisting of many machines. The idea is that each CFSM of a system is described by a simple process in the following syntax:
#!ABNF S ::= 'system' str 'of' Ptp, ..., Ptp ':' Ptp '=' M || ... || Ptp '=' M M ::= end | p ';' M | p 'do' Ptp | M '+' M | '*' M p ::= Ptp '!' str | Ptp '?' str | 'tau' str | 'tau'
A system S is given a name, specifies the (space-separated list of the) names Ptps of its machines, and has a declaration Mdec for each machine. Each name in Ptps must have a unique defining equation Ptp = M where the syntax of M is straightforward. Transition are similar those of the 'fsa' format, but they use explicit names rather than positions to address machines. Moreover, it is possible to specify internal transitions 'tau' which also carry an uninterpred annotating string. Note that branches and forks have to be surrounded by round brackets (this constraint will be relaxed in the future).
The only syntactic check made (right now) during the parsing are (i) that sender and receiver of interactions have to be different, (ii) that defining equations are unique, (iii) that communication actions name existing machines, and (iv) a machine cannot be defined as its own dual. Error messages give some information, but should be improved. Text enclosed in '[' and ']' is ignored.
Syntax of Global Graphs
The syntax of syntactic global graph is defined by the grammar:
#!ABNF G ::= Ptp '->' Ptp ':' str | G'|'G | G'+'G | G';'G | '*' G '@' Ptp | '{' G '}'
and it is essentially the one in the ICE 2016 paper where the iteration construct * G @ Ptp has been added; intuitively, such constructs repeats G until participant Ptp decides to stop the iteration.
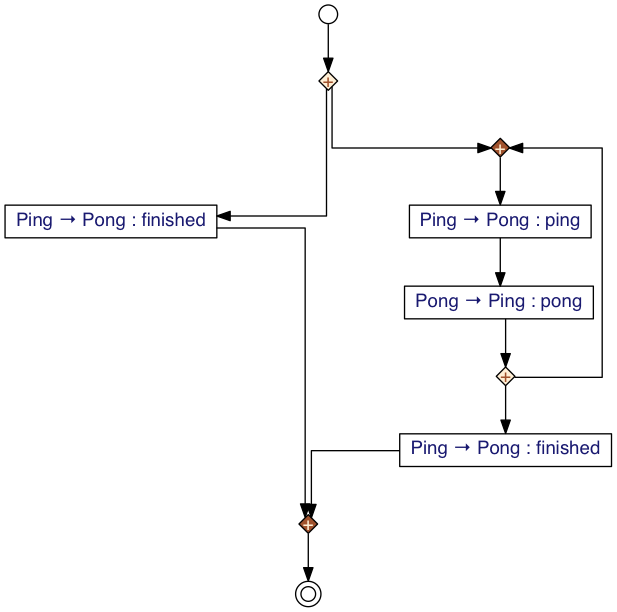
A global graph representing the ping-pong protocol in this syntax could be
#!python Ping -> Pong : finished + *{ Ping -> Pong : ping ; Pong -> Ping : pong } @ Ping ; Ping -> Pong : finished
Required Tools/ Libraries
The following tools and libraries are required.
- graphviz (and python-numpy if you would like to run the benchmarks scripts)
You can install via the command "sudo apt-get install graphviz" on ubuntu/debian
-
Haskell platform
-
MissingH (a Haskell libraries) You can install it via the command:
#! cabal install MissingH split Graphalyze
-
HKC (slight variation on git) To compile: the modified version, use the following commands:
- cd hknt-1.0/
- make (you will need OCaml, ./hknt-1.0/README)
NB: The tool has been tested on Ubuntu 13.04, 14.04, 16.04 and Mac OS X (v10.9).
Configuration
To configure chosyn, edit the assignments to 'HKCDIR' and 'PETRYDIR' in the file 'Makefile' so that the HKC and PETRY are set to the binary of hkc and petrify respectively. An example is
#!make HKCDIR = ./hknt-1.0 # change to the actual directory of hkc PETRYDIR = ../petrify/bin # change to the actual directory of petrify
Compilation
Simply
#!bash 1. make config 2. make
Running
Executing chosyn.py
The Python script 'chosyn.py' offers a command-line interface to gmc. For an overview of its usage, get the help message on the tool via the command:
#!python python chosyn.py --help
#!bash usage: chosyn.py [-h] [-v] [-df DF] [--dot DOT] [-l] [-dw DW] [-ts] [-tp TP] [-cp CP] [-p PATH] [-b BOUND] [-nc] [-pn PN] [-hkc HKC] [-gmc gmc_path] [-gg GG] [-dir DIR] [-m MUL] filename chosyn: From communicating machines to graphical choreographies positional arguments: filename Specify the path to file containing the CFSMs optional arguments: -h, --help show this help message and exit -v, --verbose Run in verbose mode -df DF Output format from dot files (svg, png, pdf, etc.) {default = svg} --dot DOT Specify the options for dot starting without '-' (e.g., --dot Nnodesep=.5) -l Suppress legend from dot files -dw DW Set fixedsize of dot nodes to the given value {default = 0} -ts Compute just the CFSMs and the transition system(s) -tp TP Pattern for colouring transitions; the syntax is "s r d msg" where s and r are the indexes of sender and receiver, d is the action, and msg is the message {default = "- - - -"} -cp CP Pattern for colouring configurations; the syntax is a string with blank-separated local state ids or '*' (as many as the number of machines) and then some blank separated string of the forom "s r" where s and r are the indexes of sender and receiver {default = ""} --path PATH Colour paths from the initial node to the ones matching the configuration pattern PATH {default: ""} -p PATH Same as --path PATH --bounded BOUND Set the bound to BOUND; if BOUND < 1, the synchronous TS is computed {default: 0} -b BOUND Same as -- bound BOUND -nc, --noclean Do not remove auxiliary files -pn PN Specify the path to petrify {default: ../petrify/bin/petrifyDarwin} -hkc HKC Specify the path to hkc {default: ./hknt-1.0/hkcDarwin} -gmc gmc_path Specify the path to gmc {default: ./gmc} -gg GG Specify the path to BuildGlobal {default: ./BuildGlobal} -dir DIR Specify the directory for the output files {default: experiments/results/}
Configuring chosyn
Besides the command line options, there are two configurations files '.chosyn.cfg' and '.dot.cfg' that can be used to tune up details. The format of the file is very basic: each line is a blank-separated pair of key / value (the lines of '.dot.cfg' also have a comment (starting with ':' after each value). You will probably just need to change the 2nd column by setting your preferred value; for instance, to change to simbol separating local states in configurations to '++' you've to change the value of 'statesep': making the line look like
#!bash 'statesep ++ :symbol to separate states in output files'
Unless you know what you're do doing, do not add lines to '.dot.cfg'. If you need to edit the file other than for altering the values as described, remember the following rules:
- do not insert text before the very first line.
- do not insert empty lines
- do not insert lines with the very same initial word
Getting the results
Upon execution, 'chosyn.py' displays some information about the files and the execution time and produces some files as results. Such files are collected in a directory that can either be a default one (currently the directory 'experiments/results/' followed by the basename of the input file) or the one you specify with the option 'dir' plus the base name of the input file. For example, if you run
#!bash python chosyn.py -nc -dw 0 -df pdf -dir experiments/results/ experiments/pingpong.fsa
chosyn: Execution Started on Sat, 15 Oct 2016 09:18:18 +0000 gmc: Parsing CFSMs file...experiments/pingpong.sys gmc: dir "experiments/results/pingpong/" gmc: Synchronous TS: (nodes 3, transitions 3) gmc: 1-bounded TS: (nodes 6, transitions 6) gmc: Branching representability: [] gmc: Branching Property (part (ii)): [] chosyn: Language-equivalence (Representability part (i))? True gg: Global graph synthesis chosyn: All done. Total execution time: 0.0250358581543 GMC check: 1.90734863281e-05 HKC minimisation: 0.00535798072815 Petrify: 0.00365400314331 Global graph generation: 0.00565099716187
on the screen and the following files are generated in the directory 'experiments/results/pingpong'
#!bash 1. pingpong_global.pdf 2. pingpong_machine_0 3. pingpong_machine_1 4. pingpong_machines.dot 5. pingpong_machines.pdf 6. pingpong_petrinet 7. pingpong_petrinet_finalpn.dot 8. pingpong_petrinet_global.dot 9. pingpong_petrinet_onesourcepn.dot 10. pingpong_petrinet_preglobal.dot 11. pingpong_projected 12. pingpong_projection_0 13. pingpong_projection_0.dot 14. pingpong_projection_0.pdf 15. pingpong_projection_1 16. pingpong_projection_1.dot 17. pingpong_projection_1.pdf 18. pingpong_toPetrify 19. pingpong_ts0.dot 20. pingpong_ts0.pdf 21. tempefc
Here is an (ugly) example of the 1-bounded transition system of the pingpong protocol generated with the command
#!bash python chosyn.py -df png -b 1 --dot "Nshape=parallelogram" --dot "Gnodesep=.5" -dir out \ -p "w *" -tp "Ping Pong * *" -cp "* * Ping Pong" experiments/pingpong.fsa

this example is meant just to show how to specify conditions on nodes and transitions using the options -p, -ts, and -cp and how to use dot options --dot (the parallelogram shape is what makes the diagram look bad). A better way to set dot preferences is to edit the file .dot.cfg described above.
Executing sgg.py
The Python script 'sgg.py' offers a command-line interface to sgg. For an overview of its usage, get the help message via the command:
#!python python sgg.py --help
#!bash usage: sgg.py [-h] [-v] [-df DF] [--dot DOT] [-l] [-dir DIR] filename sgg: semantics of syntactic global graphs and their projections to communicating machines positional arguments: filename Specify the path to file containing the CFSMs optional arguments: -h, --help show this help message and exit -v, --verbose Run in verbose mode -df DF Output format from dot files (svg, png, pdf, etc.) {default = svg} --dot DOT Options for dot starting without '-' (e.g., --dot Nnodesep=.5) -l Suppress legend from dot files -dir DIR Specify the directory for the output files {default: outputs}
Getting the results
Upon execution, 'sgg.py' displays some information about the files and produces some files as results. Such files are collected in a directory that can either be a default one (currently the directory 'experiments/results/' followed by the basename of the input file) or the one you specify with the option 'dir' plus the base name of the input file. For example, if you run
#!bash
python sgg.py experiments/examples_sgg/pingpong.sgg
sgg: start experiments/results/pingpong/in.sgg: is the initial gg experiments/results/pingpong/graph_sgg.dot: is the input gg experiments/results/pingpong/norm_sgg.dot: is the normalised initial gg experiments/results/pingpong/fact_sgg.dot: is the factorised initial gg experiments/results/pingpong/sem_sgg.dot: is the semantics of the initial gg experiments/results/pingpong/cfsmPing.dot is the machine for participant Ping experiments/results/pingpong/cfsmPong.dot is the machine for participant Pong sgg: end
Updated