Wiki
Clone wikitutorial-edinburgh2016 / CoCo / CoCo analysis of a single 1rhw simulation
Part 2: CoCo analysis of a single 1rhw simulation
Use pyCoCo to analyse all five chunks of just one of the 25 replicate simulations of 1rhw:
pyCoCo -i Edinburgh_CoCo_1rhw/rep01chunk*.nc -t Edinburgh_CoCo_1rhw/1rhw_prot.pdb -g 30 -d 2 -n 4 -s 'mass > 2.0' -o newpoints.pdb -l coco_experiment_00.log
(ve)[train62@workflow ~]$ more *.log *** pyCoCo *** Trajectory files to be analysed: ['data/rep01chunk00.nc']: frames: slice(0, 501, 1) ['data/rep01chunk01.nc']: frames: slice(0, 501, 1) ['data/rep01chunk02.nc']: frames: slice(0, 501, 1) ['data/rep01chunk03.nc']: frames: slice(0, 501, 1) ['data/rep01chunk04.nc']: frames: slice(0, 501, 1) ['data/rep01chunk05.nc']: frames: slice(0, 501, 1) Total variance in trajectory data: 2290.96 Conformational sampling map will be generated in 2 dimensions at a resolution of 30 points in each dimension. 4 complementary structures will be generated. Sampled volume: 4712.06668902 Ang.^2. Coordinates of new structures in PC space: 0 72.43 -79.45 1 30.56 -79.45 2 -48.99 32.71 3 34.75 32.71 RMSD matrix for new structures: 0.00 35.90 97.80 76.48 35.90 0.00 83.33 72.76 97.80 83.33 0.00 54.91 76.48 72.76 54.91 0.00
Total variance: This is calculated when the coordinate covariance matrix is generated. It is a measure (in units of angstrom^2) of the total variability in the (least-squares fitted) atomic coordinates of the input structures.
Sampled volume: This is the sum of the volumes of each bin in the N-dimensional histogram that is occupied by at least one data point. Note that the unit depends on the dimensionality of the analysis - as we are just using 2D in this case, the "volume" is actually an area.
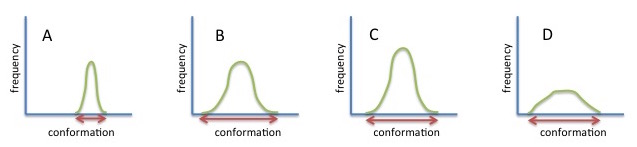
The figure below illustrates the significance of these metrics, using a 1D example of a sampling distribution. The sampling in B is better because the "volume" (a length in 1D) is longer than in A. The sampling in D is more efficient than the sampling in C - the same "volume" is sampled for a smaller number of data points; this corresponds to distribution D having the higher variance.
##Exercise 1: ##
Repeat this analysis for each 5ns chunk of the same simulation individually. Make a note of the total variance, and sampled volume, in each case.
To help make sense of this data, re-run the original pyCoCo job, analysing the whole trajectory, but add two extra arguments to the command, as shown below:
% pyCoCo -i Edinburgh_CoCo_1rhw/rep01chunk*.nc -t Edinburgh_CoCo_1rhw/1rhw_prot.pdb \ -g 30 -d 2 -n 4 -s 'mass > 2.0' -o newpoints.pdb -l coco_experiment_00.log \ --currentpoints cp.dat --newpoints np.dat
Two new files are produced: cp.dat contains the coordinates (in PC1/PC2 space) of each of the original structures. There is one line per snapshot, so 2500 lines altogether of which lines 1-500 correspond to the 500 snapshots in rep00chunk00.nc, 501-1000 are from rep00chunk01.nc, etc. The file nc.dat contains just four lines - you can probably work out for yourself what they are.
Exercise 2:
Use a graphing package of your choice to plot cp.dat and np.dat. Does the data make sense? It may help to split cp.dat into five separate files of 500 lines each (corresponding to chunk00 - chunk04) and plotting those. Consider how the graphs relate to the analysis you did in Exercise 1.
Exercise 3:
Open newpoints[0-4].pdb in a molecular viewer of your choice. Observe where CoCo has proposed new conformations for sections of the protein. On careful inspection you will probably notice that the structures appear distorted in places - this is the result of the approximation of representing structural variability by just two dimensions, and certainly hydrogen atoms will appear out of place if they were not included in the original CoCo analysis (-s 'mass > 2.0'). The impact of this on the CoCo-based enhanced sampling workflow in the ExTASY toolkit will come up later.
| << Tutorial Home | < Back | Next > |
|---|---|---|
Updated