Wiki
Clone wikiDynaMIT / Home

Welcome
Welcome to the wiki pages for DynaMIT, the Dynamic Motif Integration Toolkit!
The latest version of the toolkit is 1.1.5, released on November, 16th 2018.
DynaMIT arises from the frequent need of considering multiple tools and predictions when trying to identify true and relevant motifs in a set of biological sequences: this task is made difficult, when not completely unfeasible, by the lack of rigorous ways of integrating the results of these predictions; being able to do so, however, would greatly enhance our ability to discover robust motifs and take advantage of different prediction paradigms in the same setting.
To tackle this issue, we developed DynaMIT as a flexible, extendable and completely customizable motif integration toolkit. It is based on three phases (motif search, motif integration and results printing), each implemented by extending a standardized base component, such that anyone can develop its own motif searcher, integrator or results printer without having to deal with the toolkit internal structure: this feature allows an extreme flexiblity and adaption capability to current and future tools and strategies.
We provide many pre-implemented components out of the box, for each of the three phases: in this way, any user with common needs may use DynaMIT by simply configuring it appropriately, without having to implement any new component. Nevertheless, if one needs to use another motif searcher, design its own integration strategy, or even provide a novel way of displaying the results, it can extend the toolkit in a quick and easy way.
This wiki describes DynaMIT structure and usage in details, providing examples, common use cases and instructions on how to configure and extend the toolkit.
DynaMIT is a creation of Erik Dassi and Alessandro Quattrone:
Laboratory of Translational Genomics - CIBIO @ University of Trento
For any queries, please contact Erik Dassi (erik.dassi@unitn.it)
The paper describing the toolkit can be found here. The toolkit should be cited as follows:
- Dassi, E. and Quattrone A. (2015) DynaMIT: the dynamic motif integration toolkit. Nucl. Acids Res. doi: 10.1093/nar/gkv807.
DynaMIT workflow
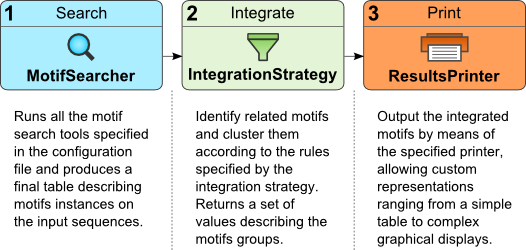
As the figure above shows, DynaMIT workflow is composed of three phases, each connected to the next one and consisting in the execution of one of the toolkit components:
-
MotifSearcher : represents the actual motif search component. By implementing this abstract class, any motif search can be run by DynaMIT, as is for the ones provided out of the box.
Results of this step are then fed to the specified integration strategies. -
IntegrationStrategy : represents the motif integration strategy component. By implementing this abstract class, a specific integration method can be applied by DynaMIT, and its results then fed to the specified results printers.
A number of strategies (Mutual information, PCA, etc.) are provided out of the box. -
ResultsPrinter: this component provides results printing capabilities. By implementing this abstract class, any kind of graphical or file output can be obtained.
Among the printers provided out of the box by DynaMIT are the simple tabular file printer, the sequence view (producing a figure indicating motifs matches on the sequences) and others.
1. Installing DynaMIT
The package can be downloaded from the Downloads page of this site or directly installed from Pypi with pip; in the first case one must unpack the downloaded archive and run:
$ python setup.py install
To install the package with pip instead, simply run:
$ pip install dynamit
To check that your installation went fine, open a Python interpreter and check that the following command produces no errors:
from dynamit import *
Both Pyhton 3.x and 2.7 are supported, although 3.6 is the suggested version.
2. The DynaMIT virtual machine
A last option for using DynaMIT consists in a virtual machine that runs Ubuntu 14.10 64-bit through the VirtualBox virtualization software. This virtual machine comes with the DynaMIT Python package pre-installed,
along with all needed software and motif search tools installed and configured. It is thus an easy way to run DynaMIT out of the box: indeed, you just need to provide the input sequences and a configuration file.
The DynaMIT virtual machine can be downloaded from here.
3. AURA 2 annotation for DynaMIT
To help in annotating sequences with known sites for post-transcriptional regulation of gene epression, we provide a set of annotations containing matrices for
RNA-binding proteins, and experimentally determined RNA-binding proteins and cis-elements sites. These can be downloaded on the AURA 2 database website: here.
4. Configuring a DynaMIT run
To specify which component of each of these categories will compose a DynaMIT run, and all related parameters, a textual configuration file is used.
Instructions on how to generate this file, either manually or through the ConfigurationGenerator, can be found in the Configuration page.
Eventually, a set of precompiled configuration files is also available in the Downloads page of this site.
5. Running DynaMIT
Once the configuration file is ready, and the sequences FASTA file to be analyzed is available, DynaMIT can be run by this simple command-line:
$ python -m dynamit <configuration_filename> <sequences filename> [-o <output_directory]
For instance:
$ python -m dynamit config.txt sequences.fasta -o myRun
The -o parameter is optional; if not specified, DynaMIT will create a directory named after the date and time of run start.
Additional optional parameters are -p followed by the number of processors to be used in the motif search phase (default = 1), and -mp followed by the minimum overlap (in nucleotides)
that each cluster motif instance must have with at least another such instance to be retained for further analysis.
Specifying this last parameter enables the motif cluster polisher, which applies such filtering with the goal of reducing false positive calls. If the parameter is not specified, this step will be skipped.
Aside from this command-line interface, DynaMIT can be integrated into existing Python code and pipelines by using its main component, the Runner.
It provides all the methods necessary to perform a complete run without the need for a command-line interface: for details on how to run DynaMIT in this way, please refer to the Runner component page.
6. Looking at the run results
Final and intermediate results will be stored into the run folder as it progresses, as described above, with filenames indicated in the console output.
In particular, the results of the first phase, motif search, will be stored in the motifSearchesResults.txt file; raw integration strategy results will instead be available in the integrationResults_<strategy name>.txt file.
Then, every ResultsPrinter specified in the configuration file will create its own folder, named results_<printer name> in which produced results files will be stored.
The filenames will be indicated in the run console output, and the description of each of these can be found in the specific ResultsPrinter page on this site.
Updated