Wiki
Clone wikicosmosis / GaussianLikelihoods
Gaussian Likelihoods in python
Many likelihoods in cosmology are (close to) Gaussian; they have the form:
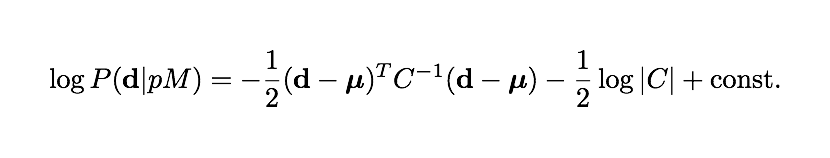
where d is a measured data vector, mu is the theory prediction for that vector, and C is the data covariance matrix, which usually comes from the noise measurements on the observations. C is often but not always a constant.
Because it is so common there is specialized CosmoSIS python code for handling Gaussian likelihoods which does many of the steps in getting log P for you.
The GaussianLikelihood class
CosmoSIS comes with a class representing general Gaussian Likelihoods, and your specific likelihood will be a subclass of that. If you're new to python classes and subclasses, read this introduction. An instance of the class will be created in the setup function (once, at the start of the program) and used in the execute function (for each point in parameter space).
You need to write a small number of methods (functions) on this class to tell it:
- what options to read from the parameter file
- how to load the data d at the start, for example load it from a file
- how to extract the theory prediction mu from the block built by the earlier pipeline steps
- how to either load the covariance C (if it is constant), or extract it from the block (if it varies)
- (optional) any cleanup to be done at the end of the pipeline
Once you have done this your new subclass can automatically generate the setup, execute, and cleanup functions you need for your module.
In the simplest cases, where the theory prediction is just an interpolation or windowed integration of a single quantity from the data block, then you just need to specify the name of the quantity to use. More generally you need to write a function telling it how to get the predicted data out of the block.
See below for four examples, vector and scalar, simple and complicated. For full details look at the file cosmosis/gaussian_likelihood.py
Simplest Vector Example
In this simplest case we just have an interpolation into a single vector quantity calculated during a pipeline. All we have to tell CosmoSIS is how to get the data and covariance, and which data columns to look in for the predicted theory observables.
from cosmosis.gaussian_likelihood import GaussianLikelihood
import numpy as np
# Note that this is a mock likelihood with very little in common with
# a real CMB data set.
class CMBLikelihood(GaussianLikelihood):
#I the simplest case, where we do not write our own
#extract_theory_points function we just look up our theory "x" and "y"
#values from the sections and names defined here.
x_section = "cmb_cl"
x_name = "ell"
y_section = "cmb_cl"
y_name = "tt"
like_name = "cmb"
def build_data(self):
#use self.options to find the data_file and load ell, tt from it
data_file = self.options.get_string("data_file", default_data_file)
ell, tt = np.loadtxt(data_file).T
return ell, tt
def build_covariance(self):
# We could load the covariance in from a file if we wanted,
# but in this mock example we will pretend we can just calculate it
# as cosmic variance only.
# We will just use these two that were saved from the outputs of build_data
ell = self.data_x
c_ell = self.data_y
#We have to return a matrix, though in this case it is just
#a diagonal one. Here is the diagonal
sigma2 = 2 * c_ell**2 / (2*ell+1)
#convert into matrix and return
covmat = np.diag(sigma2)
return covmat
#The Gaussian likelihood makes the setup, execute, and cleanup functions for us
#in this case
setup, execute, cleanup = CMBLikelihood.build_module()
Complicated Vector Example
Here's an example of a more complicated BAO likelihood. In this example we over-ride the function that pulls the predicted observable out of the data block:
from cosmosis.gaussian_likelihood import GaussianLikelihood
from cosmosis.datablock import names
import os
import numpy as np
dirname = os.path.split(__file__)[0]
default_data_file = os.path.join(dirname, "wigglez_bao.txt")
default_weight_file = os.path.join(dirname, "wigglez_bao_weight.txt")
default_rs_fiducial = 148.6
class WigglezBAOLikelihood(GaussianLikelihood):
like_name = "wigglez"
def __init__(self, options):
#This calls the parent class's init function first
super(WigglezBAOLikelihood, self).__init__(options)
#Now we have some of our own additional options to save:
#We will need the fiducial last-scattering sound horizon, so read it in here.
#The self.options object contains all the options from the section
#of the ini file describing this module
self.rs_fiducial = self.options.get_double("rs_fiducial", default_rs_fiducial)
#And a verbosity setting
self.verbose = self.options.get_bool("verbose", False)
def build_data(self):
#use self.options to
data_file = self.options.get_string("data_file", default_data_file)
z, Dv = np.loadtxt(data_file).T
return z, Dv
def build_covariance(self):
weight_file = self.options.get_string("weight_file", default_weight_file)
weight_matrix = np.loadtxt(weight_file)
#Also save this; see note below in build_inverse_covariance
self.inv_cov = weight_matrix
covariance = np.linalg.inv(weight_matrix)
return covariance
def build_inverse_covariance(self):
#This likelihood is a little different than most in that we load
#the weight matrix from file rather than its inverse, the covariance.
#so we have to work around the default order things are loaded in
#the parent class by doing this
return self.inv_cov
def extract_theory_points(self, block):
#load distance relations and R_S
z = block[names.distances, "z"]
da = block[names.distances, "D_A"] # in Mpc
H = block[names.distances, "H"] # in c/Mpc
rs = block[names.distances, "RS_ZDRAG"]
#Compute the derived D_V distance
dr = z / H #in Mpc/c
dv = (da**2 * (1+z)**2 * dr)**(1./3.)
# z and dv are loaded in chronological order;
# reverse to start from low z
z = z[::-1]
dv = dv[::-1]
#Interpolate into the theory at the
#observed redshifts
z_data = self.data_x
dv_predicted = np.interp(z_data, z, dv)
#If required, print out some info
if self.verbose:
print "dv_predicted = ", dv_predicted
print "dv_data = ", self.data_y
print "rs = ", rs
print "rs_fiducial = ", self.rs_fiducial
#Return the quantity correctly scaled for the sound horizon
return dv_predicted*self.rs_fiducial/rs
setup, execute, cleanup = WigglezBAOLikelihood.build_module()
Simplest Scalar Example
In the simplest case there is just a single parameter whose likelihood want, which we can get from a single location in the data block (i.e. it has already been calculated in the pipeline). In this code the user can also override the mean, sigma, section, and parameter name on the command line if they want to:
from cosmosis.datablock import names
from cosmosis.gaussian_likelihood import SingleValueGaussianLikelihood
class Riess11Likelihood(SingleValueGaussianLikelihood):
# The mean and standard deviation of the Riess11 measurements.
# The user can over-ride these in the parameter file if desired
mean = 0.738
sigma = 0.024
#The value (either chosen by the sampler or computed
#somewhere in the pipeline) that this Likelihood uses.
#If we needed to operate on a derived quantity we could
#have redefined the method block_theory_points instead; see below
section = names.cosmological_parameters
name = "h0"
#The name we should use for the likelihood
like_name = "riess"
setup, execute, cleanup = Riess11Likelihood.build_module()
More Complicated Scalar Example
Our pipeline might not have previously calculated/created the particular value we need in the likelihood previously, as it did in the last example. We might need to calculate it in the likelihood module.
In this case we override a method, extract_theory_points, to load the theory points that we need:
from cosmosis.datablock import names
from cosmosis.gaussian_likelihood import SingleValueGaussianLikelihood
class BBNLikelihood(SingleValueGaussianLikelihood):
# The mean and standard deviation of the BBN measurements.
# The user can over-ride these in the ini file if desired
mean = 0.023
sigma = 0.002
#We will use this section ourselves in the method below this time
section = names.cosmological_parameters
#The name of the likelihood
like_name = "bbn"
# This method is the new bit - here we compute and return the value we need,
# in this case the combination omega_b * h**2
def extract_theory_points(self, block):
omega_b = block[self.section, 'omega_b']
h = block[self.section, 'h0']
ombh2 = omega_b * h**2
return ombh2
setup, execute, cleanup = BBNLikelihood.build_module()
Updated