Thank you very much for your report.
The current version of LipidHunter was tested with mzML files converted by ProteoWizard from Waters and Thermo raw files.
What we have observed is that the mzML files converted from different vendors have different structure in the data storage part.
As a result, vendor specific algorithms were applied in several sub-processes during processing.
We have a wiki page to address this mzML file format issue, please have a look: LipidHunter.mzML format requirements.
It would be nice if you can send us a mzML file you converted, so we can have a look and add support to Agilent .d files.
Please try to convert 1-2 min with MS2 level from your raw data and send it to me by email( zhixu.ni<at>uni-leipzig.de).
Thank you!
Zhixu Ni
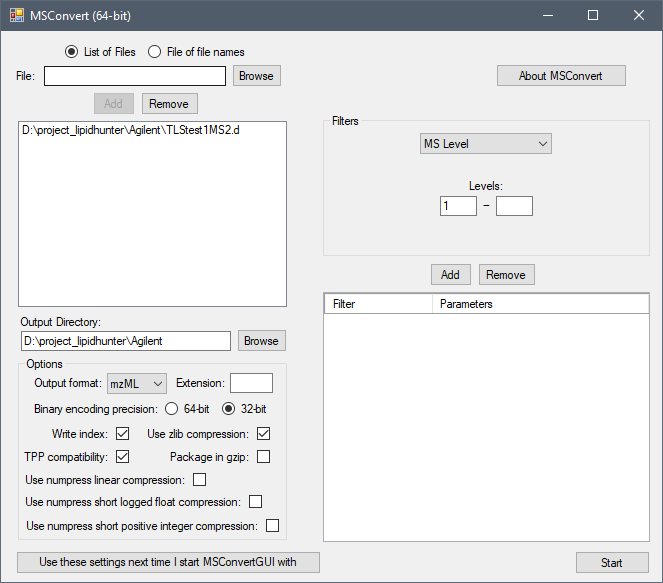

Thank you for the sample data.
I had a look of
Agilent.d raw data.Please use following parameters to convert your data:
The converted
mzMLfile with MS2 information is usually at least 300MB and you can use the file for all steps from Step I to Step V.Please make sure you selected
Thermomode before staring any step formzMLfromAgilent.We recommend you to use +/- 0.05 m/z for LIPID MAPS bulk search in your first run. You can lower the tolerance base on your instrument resolution step by step in the later runs.