Hi!
Thank you for using LipidHunter! The current version we did not check the compatibility of mzML converted from Sciex data. If you have some sample data file using DDA, please try to convert to mzML and send me both raw files and corresponding mzML to us. please also provide us the screenshot of the MSConvert with your conversion settings. If you would like to share your files to us, please put the files to Google Drive, One Drive, other online storage so we can have a look.
Thank you for choosing LipidHunter for your Lipidomics analysis!
Best wishes,
Zhixu Ni
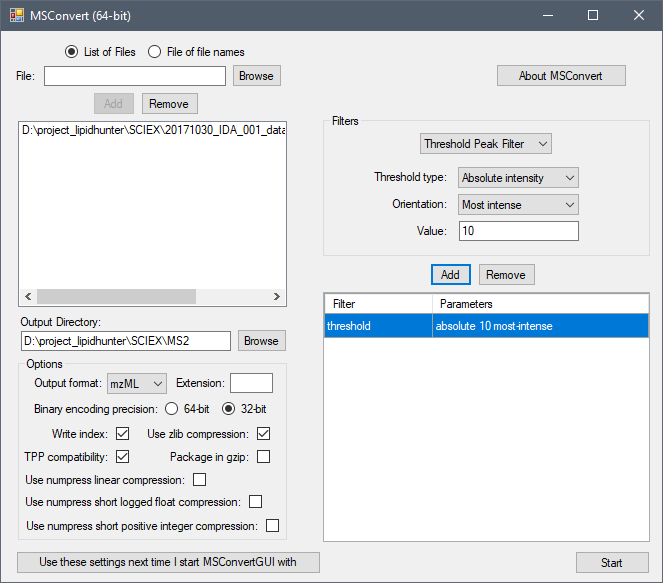
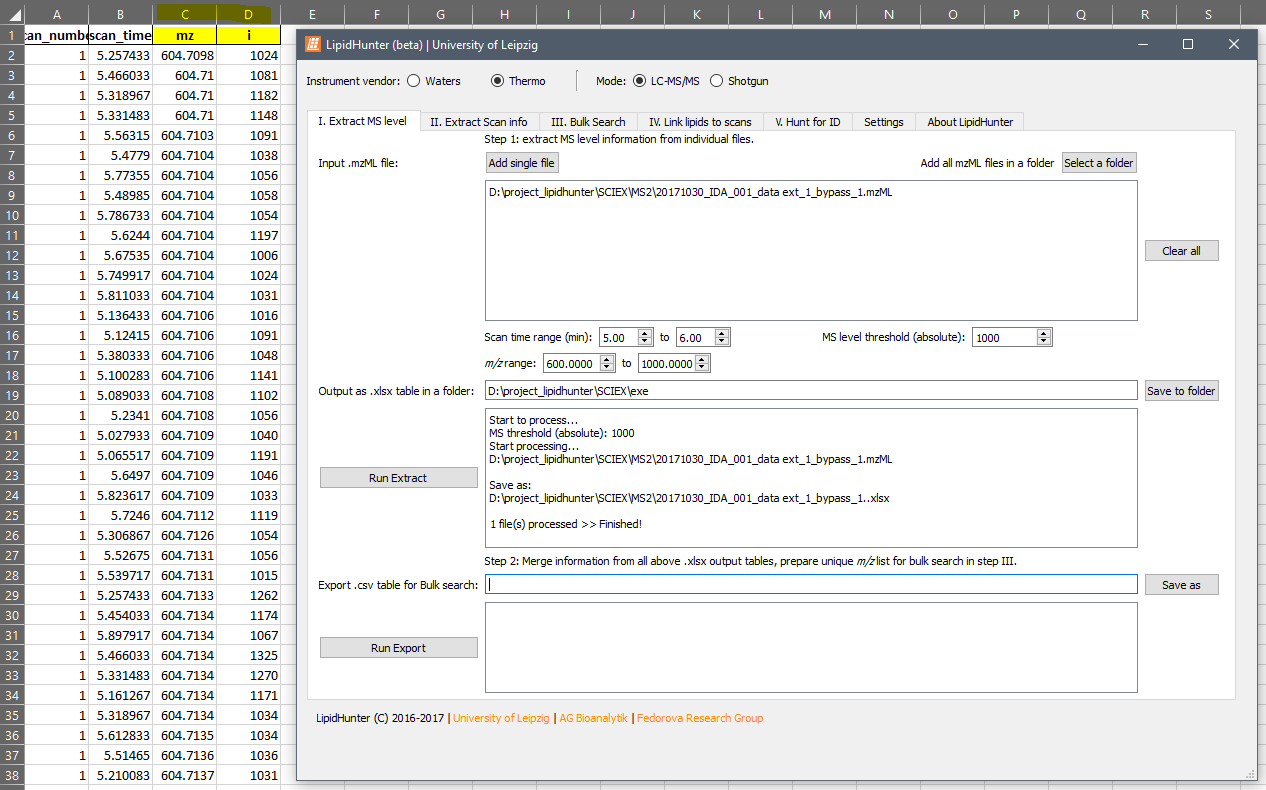
 Here are my settings
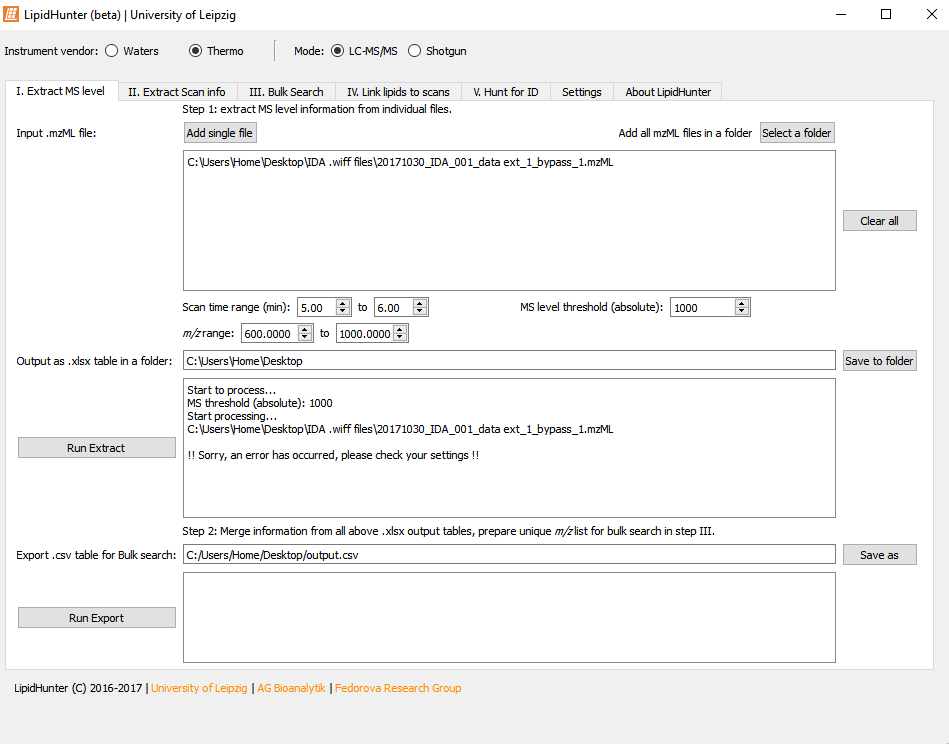
Here are my settings  I just left it at the default setting. Apologies as I am very new to this.
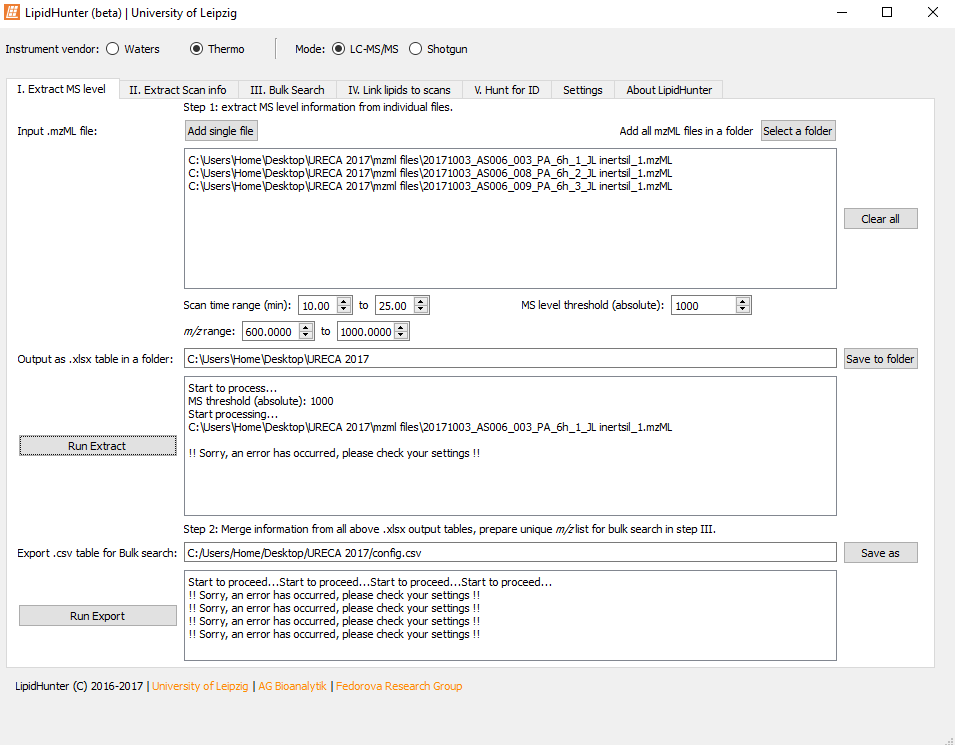
Really appreciate if you could let me know why I am getting this error.
I just left it at the default setting. Apologies as I am very new to this.
Really appreciate if you could let me know why I am getting this error.

Hi Pandiyan!
It's good to know that the file finally works for you!
The problem is you cannot download version 3.0.9134 from ProteoWizard project website any more, and I am not sure if I am allowed to send you my installed version.
The alternative way is to download version 3.0.10158 from ProteoWizard project website:
http://teamcity.labkey.org:8080/viewLog.html?buildId=380712&buildTypeId=bt83&tab=artifacts
I suggest you to uninstall your old ProteoWizard, restart your computer and then install the following version:
http://teamcity.labkey.org:8080/repository/download/bt83/380712:id/pwiz-setup-3.0.10158-x86_64.msi
I have already tested this version of ProteoWizard, it was working fine and the converted .mzML file from your .wiff data is compatible with LipidHunter.
Please let me know if you succeed with ProteoWizard version 3.0.10158 or not.
If this version works, please keep the installation file for your later projects.
Best wishes,
Zhixu Ni