Wiki
Clone wikitutorial-edinburgh2016 / CoCo / Analysing MD Data with CoCo
Aim of the tutorial.
In this tutorial you will see how CoCo can be used to explore the characteristics of ensembles of protein structures generated by MD simulation. Before you start you will need:
- The tutorial data - supplied for you but also available here.
- The ExTASY tool pyCoCo installed - done for you, but also available for download here.
- Access to a simple graph drawing package (e.g. gnuplot).
- Access to a molecular visualisation tool - e.g. VMD/Chimera/pyMol.
What is CoCo?
CoCo ("Complementary Coordinates") is a method for testing and potentially enriching the the variety of conformations within an ensemble of molecular structures. It was originally developed with NMR datasets in mind and the background and this application is described in:
CoCo, which is based on principal component analysis, analyses the distribution of an ensemble of structures in conformational space, and generates a new ensemble that fills gaps in the distribution. These new structures are not guaranteed to be valid members of the ensemble, but should be treated as possible, approximate, new solutions for refinement against the original data. Though developed with protein NMR data in mind, the method is quite general – the initial structures do not have to come from NMR data, and can be of nucleic acids, carbohydrates, etc.
The outline of the CoCo method is as follows:
- Step 1: The existing ensemble is analysed by PCA and the distribution of the snapshots in a low-dimensional PC space determined:
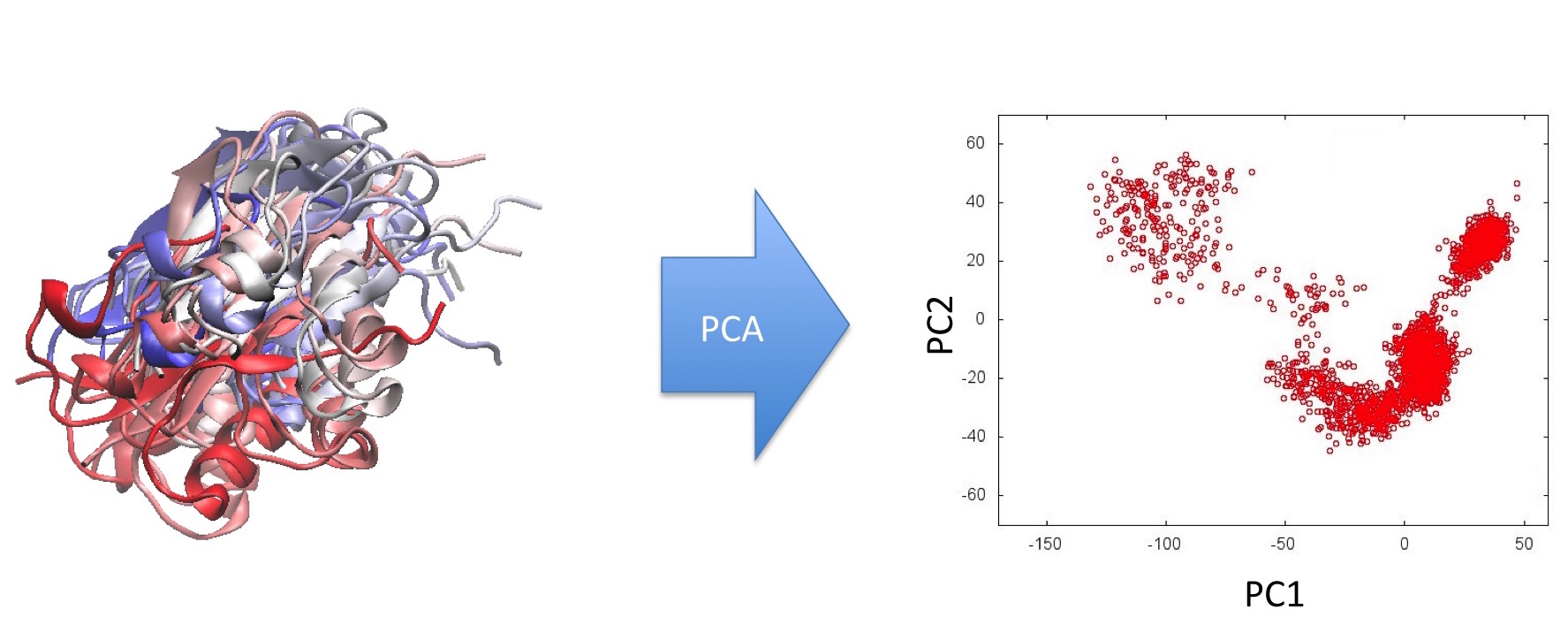
Step 2: The CoCo process is used to identify so-far unsampled regions of this PC subspace: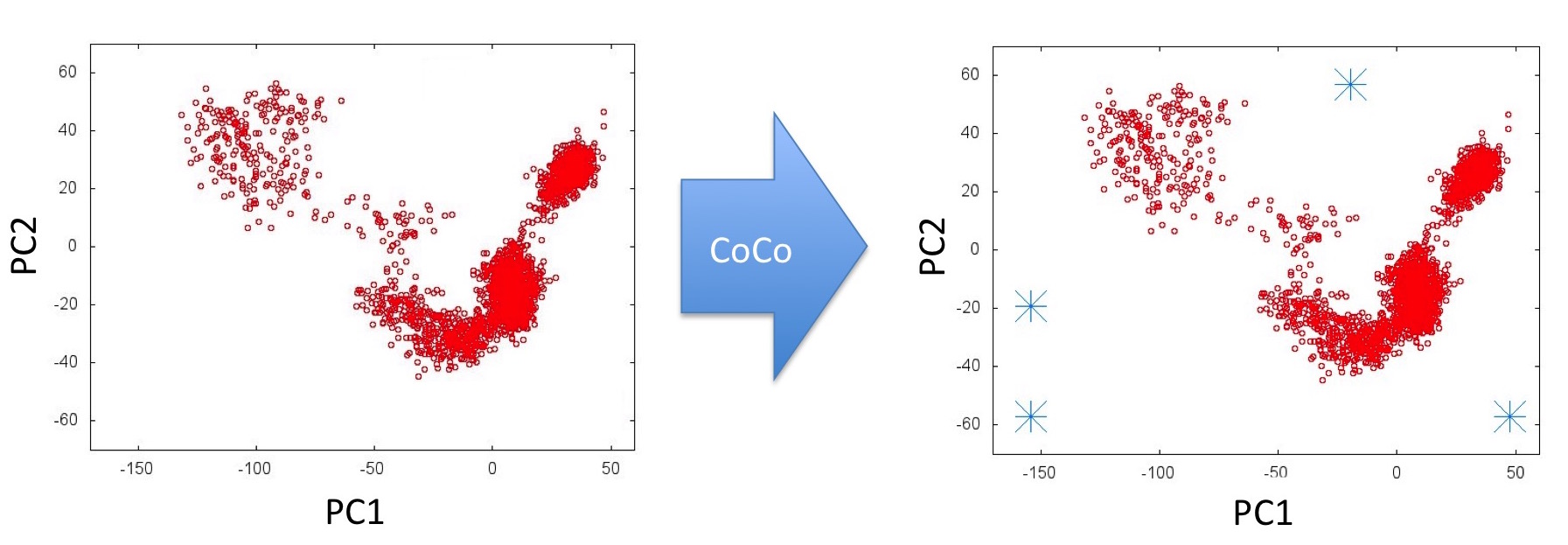
Step 3: The CoCo process generates candidate structures for the molecule corresponding to the unsampled points: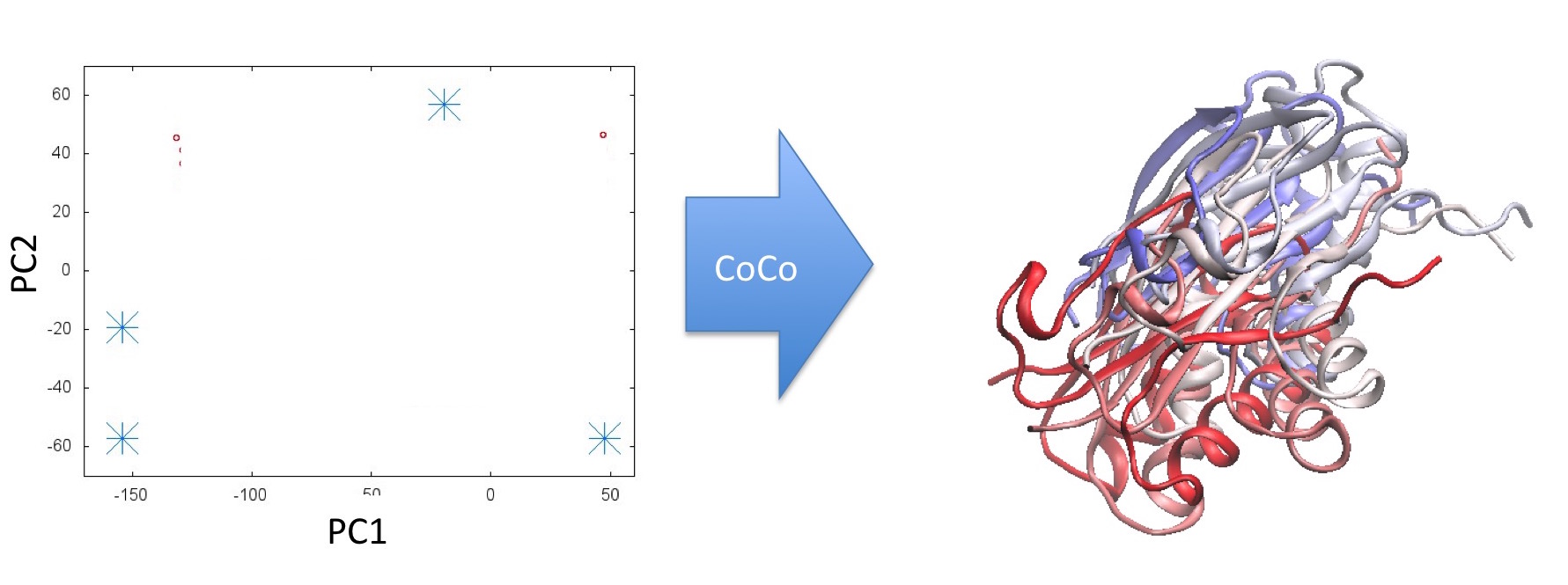
The data you will analyse.
In the folder ./Edinburgh_CoCo_1rhw/ are a set of MD trajectory files for a small protein - dynein light chain LC8 (PDB code 1rhw). Twenty five replicate 25 ns simulations of this (rep01 - rep25) have been run using Amber. Each trajectory file, stripped of water, has been split into 5ns chunks (chunk00 - chunk04). Also in this folder is a pdb format file for the protein (1rhw_prot.pdb).
% ls data 1rhw_prot.pdb rep07chunk01.nc rep13chunk03.nc rep20chunk00.nc rep01chunk00.nc rep07chunk02.nc rep13chunk04.nc rep20chunk01.nc rep01chunk01.nc rep07chunk03.nc rep14chunk00.nc rep20chunk02.nc rep01chunk02.nc rep07chunk04.nc rep14chunk01.nc rep20chunk03.nc rep01chunk03.nc rep08chunk00.nc rep14chunk02.nc rep20chunk04.nc ... rep06chunk01.nc rep12chunk03.nc rep19chunk00.nc rep25chunk02.nc rep06chunk02.nc rep12chunk04.nc rep19chunk01.nc rep25chunk03.nc rep06chunk03.nc rep13chunk00.nc rep19chunk02.nc rep25chunk04.nc rep06chunk04.nc rep13chunk01.nc rep19chunk03.nc rep07chunk00.nc rep13chunk02.nc rep19chunk04.nc
Part 1: Introduction to pyCoCo.
The ExTASY tool pyCoCo will be used for the analysis. First just check your installation is working OK:
% pyCoCo -h usage: pyCoCo [-h] [-g GRID] [-d DIMS] [-n FRONTPOINTS] -i [MDFILE [MDFILE ...]] -o OUTPUT -t TOPFILE [-v] [-l LOGFILE] [-s SELECTION] [--nompi] [-V] [-f FMT] [--currentpoints CURRENTPOINTS] [--newpoints NEWPOINTS] optional arguments: -h, --help show this help message and exit -g GRID, --grid GRID Number of points along each dimension of the CoCo histogram -d DIMS, --dims DIMS The number of projections to consider from the input pcz file in CoCo; this will also correspond to the number of dimensions of the histogram. -n FRONTPOINTS, --frontpoints FRONTPOINTS The number of new frontier points to select through CoCo. -i [MDFILE [MDFILE ...]], --mdfile [MDFILE [MDFILE ...]] The MD files to process. -o OUTPUT, --output OUTPUT Basename of the pdb files that will be produced. -t TOPFILE, --topfile TOPFILE Topology file. -v, --verbosity Increase output verbosity. -l LOGFILE, --logfile LOGFILE Optional log file. -s SELECTION, --selection SELECTION Optional atom selection string. --nompi Disables any attempt to use MPI. -V, --version show program's version number and exit -f FMT, --fmt FMT Optional output format. --currentpoints CURRENTPOINTS Optional file with coordinates of current points. --newpoints NEWPOINTS Optional file with coordinates of CoCo-generated points.
-g GRID:
The CoCo method generates a multi-dimensional histogram of the ensemble data in the PC subspace. The -g option (e.g. -g 20) is used to define how many bins will be used per dimension. If not specified, 10 bins are used:
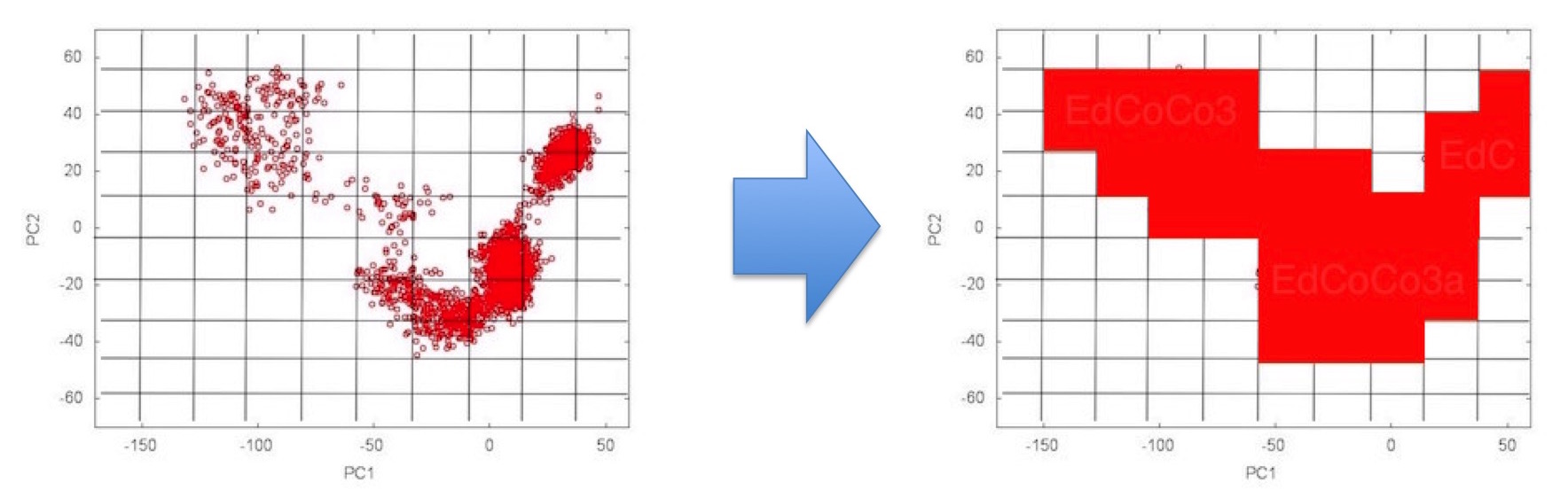
-d DIMS: CoCo histograms are typically three or four dimensional (rather than the 2D maps shown here to demonstrate the principles), the choice is made here (e.g. -d 4). If not specified a 3D histogram (PC1/PC2/PC3) is used.
-n FRONTPOINTS: This sets the number of new conformations, in so-far unsampled regions of the PC map, will be generated by the CoCo process. If not specified, just one new point is produced (equivalent to -d 1).
-o OUTPUT: This defines the names of the files with the new structures. So '-o newpoints.pdb' will produce files newpoints1.pdb, newpoints2.pdb, newpoints3.pdb ... up to the number FRONTPOINTS. Files can be written in three formats, identified by the file extension: .pdb, .gro (Gromacs) or .rst7 (Amber). If you have a non-standard extension name, you can use the -f option to tell pyCoCo what format to write.
-i MDFILE: pyCoCo accepts MD files in a range of common formats (.xtc, .nc, .dcd, etc.), and multiple files can be specified as well (e.g. -i traj1.dcd traj2.xtc traj3.xtc). The only limitation is that all must be compatible with the topology file (see below) - i.e, have the same number of atoms, in the same order.
-t TOPFILE: A topology file. Acceptable formats are .pdb or .gro.
-l LOGFILE: An output file with details of the CoCo analysis. More comprehensive that the messages written to the screen when the '-v' flag is used.
-s SELECTION: You can select which atoms from the trajectory file to use in the CoCo analysis. If this option is not given all atoms are used. The syntax for this comes from the underlying MDTraj library: see here for details.
We will cover some of the other options a bit later.
| <<Tutorial Home | Next > | |
|---|---|---|
Updated