Wiki
Clone wikienmap-box-idl / synthMix-SVR Manual for Application
Concept
synthMix-SVR is an IDL-based tool for quantitative analysis of remote sensing data. It implements the concept of combining support vector regression (SVR) with synthetically mixed training data for mapping sub-pixel fractions of land cover (Okujeni et al. 2013). synthMix-SVR is embedded into the EnMAP-Box 2.1 (available from: www.enmap.org) and makes use of imageSVM 3.0 (available from: www.imagesvm.net) for SVR modeling.
The goal of synthMix-SVR is to provide a user-friendly tool for producing land cover fraction maps from remote sensing imagery. synthMix-SVR follows the approach proposed in Okujeni et al. (2013):
- A spectral library is used to generate a synthetically mixed training data set for a single land cover category of interest, a so-called target category.
- The synthetic data set is used to train a SVR model, which is subsequently used to derive a fraction map of the respective target category.
Building on this concept, synthMix-SVR combines (1) and (2) and additionally allows the user to flexibly map multiple target categories through iterative processing (Figure 1). Thus, synthMix-SVR is suited for a comprehensive mapping of land cover, delivering a set of multiple target category fraction maps in a multilayer image.
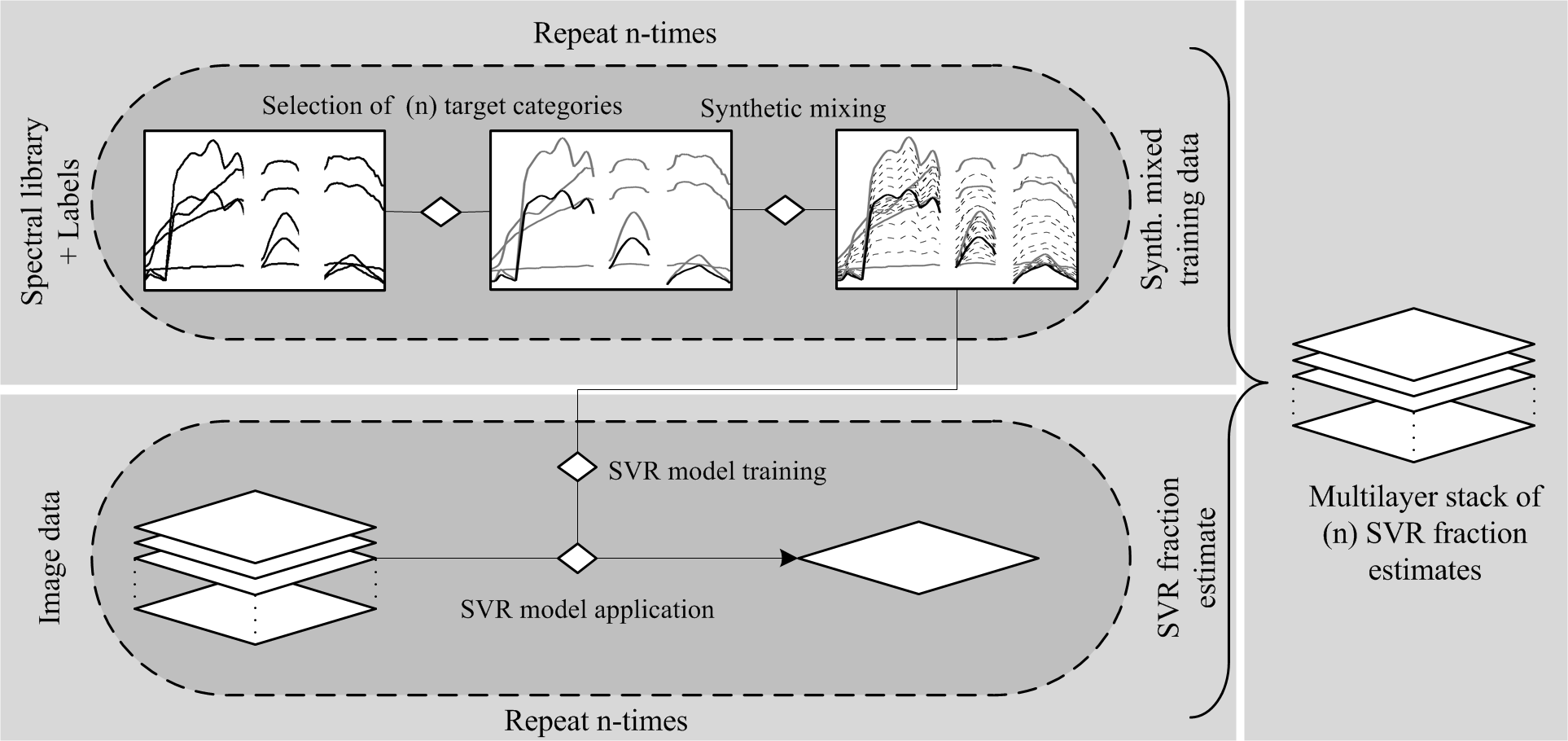
Figure 1 Workflow for quantifying land cover using synthMix-SVR (Okujeni et al. (2013), modified).
synthMix-SVR is developed as a open source product at the Geomatics Lab of Humboldt-Universität zu Berlin. By distributing the tool, the authors hope to enlarge the number of applications and in this way learn more about its potential and weakness.
synthMix-SVR uses generic file formats (ENVI file types) as used by the EnMAP-Box 2.0. These formats are used for spectral image data as well as spectral libraries and library labels. The labeling of library spectra is not supported by synthMix-SVR and has to be carried out using the Labeling Tool of the EnMAP-Box.
The synthMix-SVR application is distributed with a test data set, consisting of a hyperspectral image subset and a corresponding spectral library. Once the data have been loaded into the EnMAP-Box, synthMix-SVR guides the user through several processing steps. This includes (i) management of input and output data, (ii) selection of target categories, (iii) definition of mixing parameters during the training data generation, and (iv) an automatized or user-defined SVR model parameterization with subsequent model application. Further optional features of synthMix-SVR enable the superposition of random noise on spectra and image post-processing to produce physically meaningful fraction maps.
Background
Support vector machines (SVMs) emanate from the field of machine learning and provide flexible, non-parametric and nonlinear models that are excellently suited for exploiting remote sensing image data (e.g. Foody and Mathur 2004; Melgani and Bruzzone 2004; Camps-Valls et al. 2006; Durbha et al. 2007). Detailed introductions into support vector machines and their underlying concepts can be found in Schölkopf and Smola (2002), Smola and Schölkopf (2004) or Burges (1998). While the support vector classifier (SVC) has been established as a powerful technique for the per-pixel mapping of discrete land cover classes, little attention has been paid to the use of support vector regression (SVR) for estimating sub-pixel fractions of land cover. This can be partially explained by the difficulty in finding reliable quantitative training information, i.e., pairs of spectra and associated cover fraction, needed for regression modeling. Compared to per-pixel classifiers, training signatures can hardly be labeled in the data itself or mapped in the field. A further possible solution to combine image spectra with spatially aggregated land cover information from a high resolution reference map often fails due to inaccurate co-registered data sets. In this context, the combination of SVR with synthetically mixed training data has been demonstrated to overcome this drawback and was therefore recommended as suitable approach for sub-pixel mapping purposes (Okujeni et al. 2013).
Generation of synthetically mixed training data
The general idea behind generating synthetically mixed training data is to produce a set of multiple mixed spectra along with related mixing fractions, which can be used as training input for regression modeling of a single target land cover category (Figure 2). A library consisting of pure material spectra that are assigned to their land cover category forms the data base. Following the description in Okujeni et al. (2013), further processing steps include:
- The partitioning of the library spectra into a target category and a background category (includes all remaining categories).
- The calculation of synthetic mixtures between each pure spectrum of the target category (with 100% mixing fraction) and each pure spectrum of the background category (with 0% mixing fraction). For simplification, linear mixing systematics is assumed. Further, the user needs to define the mixing parameters, including the mixing complexity (number of possible material spectra to be mixed) and the mixing interval (number of intermediate mixtures within the fraction range between 0 and 100%).
- The combination of all pure original and mixed spectra in a single spectral library. The mixing fraction of the respective target category is assigned to each spectrum.
synthMix-SVR was developed to generate synthetically mixed training data for multiple target categories through iterative processing. The user is requested to set the mixing interval. The current version of synthMix-SVR only supports the generation of binary mixtures between each pure spectrum of the target category and each pure spectrum of the background category. To account for environmental or instrumental errors, the user may optionally add noise to the spectral data.
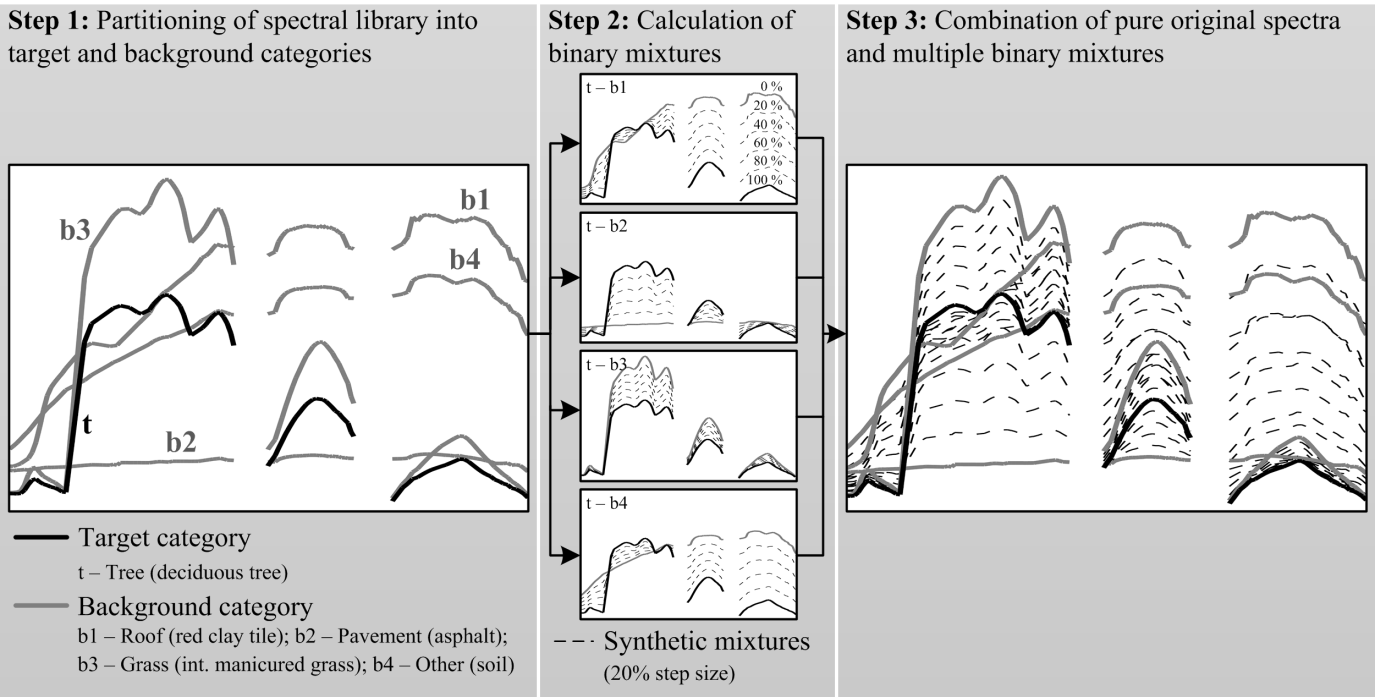
Figure 2 Generation of synthetically mixed training data (Okujeni et al. (2013)).
Support vector regression modeling
SVR has been widely used as powerful, nonlinear technique mainly for quantifying biophysical/-chemical plant properties (Camps-Valls et al. 2006; Durbha et al. 2007; Tuia et al. 2011). In general, SVR estimates a linear dependency between pairs of n-dimensional input vectors (i.e., spectral bands) and a 1-dimensional target variable (i.e., land cover fraction of a target category) by fitting an optimal approximating hyperplane to the training data. For nonlinear problems, the training data are implicitly mapped by a kernel function into a higher dimensional space, wherein the new data distribution enables a better fitting of a linear hyperplane. The parameterization of an SVR requires the user to select the parameter(s) of a kernel function g as well as the regularization C and loss function ε parameters. Once these parameters have been selected, the optimal approximating hyperplane is found by quadratic optimization.
synthMix-SVR integrates the SVR algorithm provided by imageSVM 3.0 (available from: www.imagesvm.net). imageSVM is an IDL based tool for the SVM classification and regression analysis of remote sensing image data. imageSVM uses LIBSVM (Chang and Lin 2011) and a Gaussian kernel function during the training of the SVM. synthMix-SVR makes use of the imageSVM graphical user interface, which enables (i) the automatized or user-defined SVR model parameterization via grid search and internal validation, and (ii) the subsequent model application to derive a model prediction. Once the synthetically mixed data for the selected target categories have been generated, synthMix-SVR iteratively trains SVR models and derives fraction maps through model application to the image data.
Post-processing of fraction maps
Land cover fractions predicted by SVR cover continuous, physically meaningful fraction values between 0 and 100% through partial interpolations of the training data interval. However, improper extrapolations may also result in unrealistic fractions, i.e., negative values (below 0%) or super-positive values (greater than 100%). Beyond, through mapping single land cover categories independently from each other, it cannot be guaranteed that the combination of all fraction maps sum to unity (100%). A comprehensive analysis and discussion is provided in Okujeni et al. (2013).
The synthMix-SVR post-processing module was designed to optionally account for unrealistic fraction values and to produce meaningful stacks of fraction maps that sum to unity.
User guide
Data requirements and file formats
Fraction mapping using synthMix-SVR requires a spectral image, a spectral library and a classification label file that is related to the library spectra:
- Image data are expected to be stored as ENVI Standard file. The EnMAP-Box provides functionalities for importing other file types (e.g. TIFF, ASCII).
- Spectral libraries are expected to be stored as ENVI Spectral Library file. The EnMAP-Box provides functionalities for importing other file types (e.g. ASD file, ASCII, CSV-tables).
- The classification label file is a pseudo image stored as ENVI Classification file and contains the class information of the library spectra. After importing the spectral library into the EnMAP-Box, the Labeling Tool can be used to create a corresponding classification label file.
For further information, see the EnMAP-Box Manual or Data Format Definition.
Getting started
After starting the EnMAP-Box 2.1, a test data set can be opened from the EnMAP-Box File Menu. It includes a subset of a hyperspectral urban scene of Berlin, Germany, (HyMap data, 9 m spatial resolution, 111 spectral bands) and a spectral library with 41 pure image spectra assigned to the four categories impervious (23 spectra), grass (5), tree (7) and other (6). More detailed information on the data set can be found in Okujeni et al. (2013) and Okujeni et al. (2014).
- Select Applications > Unmixing > synthMix-SVR > Load Test Data. The test data set will appear in the EnMAP-Box Filelist.
- Use the drag-and-drop functionality to display the test data set in the EnMAP-Box View Manager.
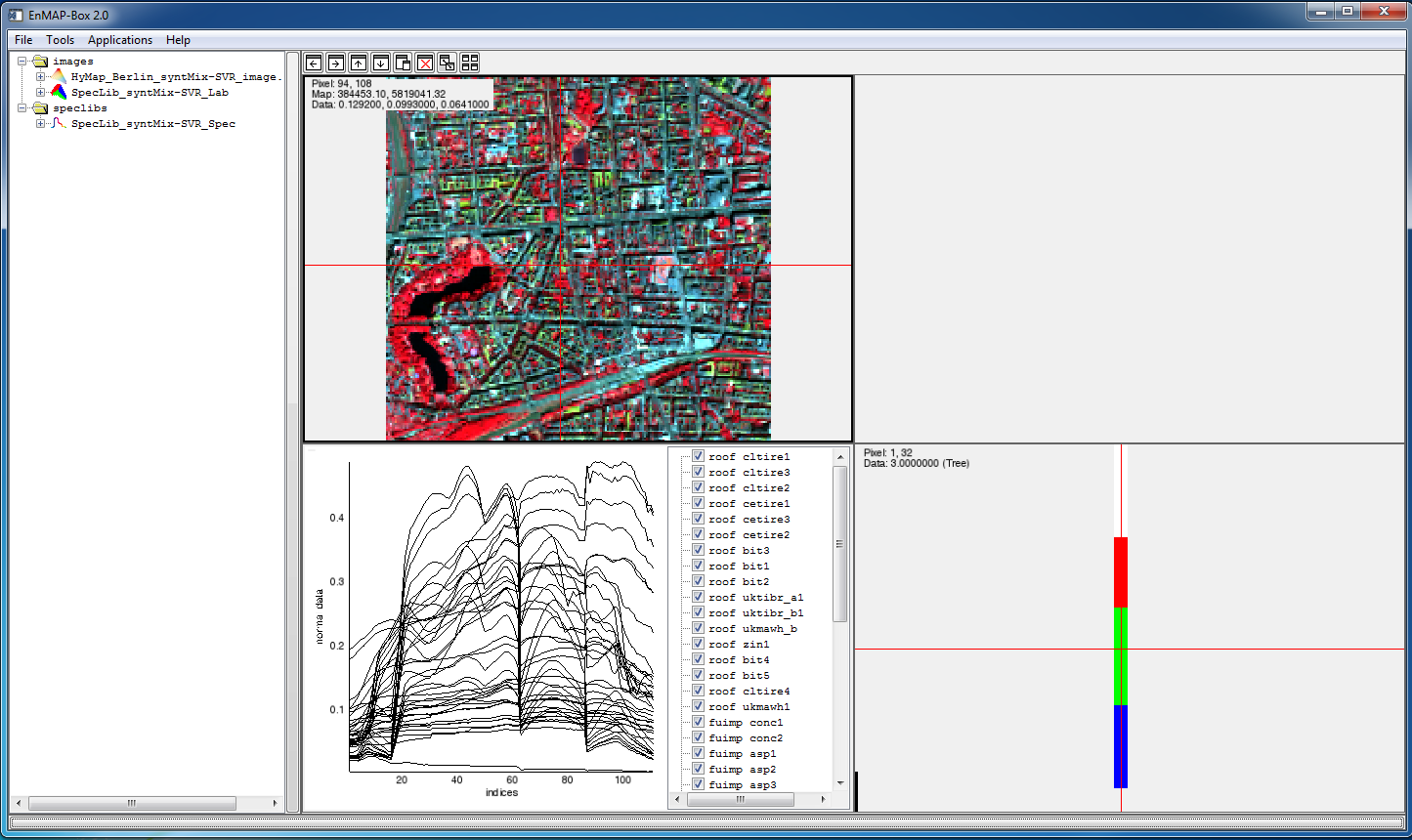
Run synthMix-SVR
Open synthMix-SVR
After loading the test data set, synthMix-SVR can be started from EnMAP-Box Application Menu.
- Select Applications > Unmixing > synthMix-SVR > synthMix-SVR.
Manage data input and output
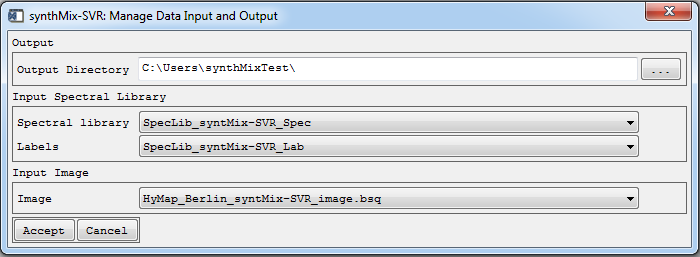
In the first dialog, the user is asked to specify the input data (spectral library, classification label and image data) and the output data folder where all produced data sets (training data, SVR models and fraction maps) will be stored.
- Specify the settings (as shown above) and click Accept to proceed.
Generate synthetically mixed training data
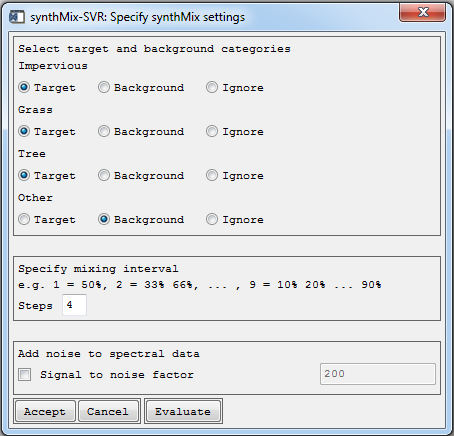
During the generation of synthetic mixtures, the user is requested specify settings for generation synthetically mixed training data:
- Select target and background categories: Displays the land cover categories according to the classification label file. The user is asked to specify the target categories (synthetic training data, SVR models and land cover fraction maps will be produced) or categories which will only be used in the background during the generation of synthetic mixtures. The ignore option may be used to exclude categories entirely from the analysis. The user must choose at least one target and one background category or two target categories.
- Specify mixing interval: The mixing interval refers to the number of intermediate mixtures between 0 and 100% when mixing target category spectra (with mixing fraction of 100%) against background category spectra (with mixing fraction of 0%). The mixing interval should balance the trade-off between necessary and avoidable number of mixtures, considering accuracy of fraction maps and computing time. In synthMix-SVR, the mixing interval is defined equally for all land cover categories and is specified via mixing steps:
| Mixing step | Interval width | Output mixing fractions of target category |
|---|---|---|
| 1 | 50% | 0%, 50%, 100% |
| 2 | 33% | 0%, 33%, 66%, 100% |
| 3 | 25% | 0%, 20%, 40%, 60%, 89%, 100% |
| 4 | 20% | 0%, 25%, 50%, 75%, 100% |
| … | … | … |
| 9 | 10% | 0%, 10%, 20%, 30%, 40%, 50%, … 90%, 100% |
- Add noise to spectral data: Optionally, pure and synthetically mixed spectra can be imposed by noise to account for environmental or instrumental errors. The noise-degraded signal
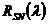 is calculated according to Okin et al. (2001):
is calculated according to Okin et al. (2001):
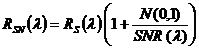 , where,
, where, 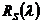 is the noise-free spectrum,
is the noise-free spectrum, 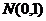 is a random number generated from a normal distribution with a mean of 0 and standard deviation of 1 and
is a random number generated from a normal distribution with a mean of 0 and standard deviation of 1 and 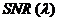 is the signal-to-noise ratio.
is the signal-to-noise ratio.
- Enter the settings as shown above.
Before accepting the dialog, the user may evaluate the settings in order to get an overview of how many synthetically mixed training data sets and spectra are created. Note that the SVR processing time increases with increasing input data.
- Click Evaluate.
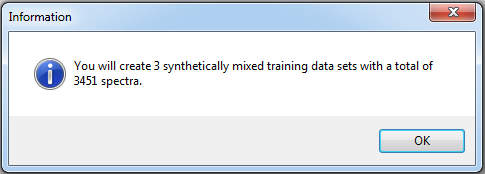
- Click OK to get back to previous dialog. Adjust settings if necessary and click Accept to proceed.
The generated synthetically mixed training data sets, i.e., pure original and synthetically mixed spectra as well as related mixing fractions, for the selected target categories will be created, saved in the defined output folder and opened in the EnMAP-Box File Menu.
Specify post-processing
The following dialog allows the user to optionally specify post-processing options that will be carried out once the fraction maps are produced.
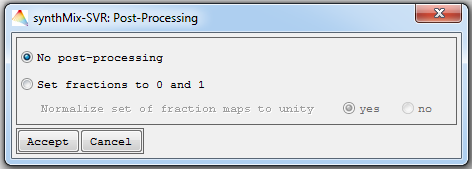
No post-processing: Fraction maps will not be post-processed.
Set fractions to 0 and 1: Negative (below 0%) and super-positive (above 100%) fraction values will be set to 0 and 100%.
Normalize set of fraction maps: Weights the estimated fractions of a target category relative to the sum of estimated fractions of all target categories on a per-pixel basis. This way all target category fraction maps sum to unity (i.e., 100%).
- Specify settings and click Accept to proceed.
Note that all files, including original and the post-processed fraction maps will be saved in the output folder when post-processing options are selected.
SVR modeling
SVR modeling using synthMix-SVR is based on imageSVM, which allows the user to parameterize SVR models based on default values or advanced settings prior to applying the model to the image data. SVR parameters g and C, depend on the data range and distribution and are thus case specific. A common strategy to search for adequate values for g and C is a two-dimensional grid search with internal validation. This strategy is implemented in imageSVM. For tuning the loss function parameter ε we use a efficient search heuristic proposed by Rabe et al. (2013)
SVR modeling using default values
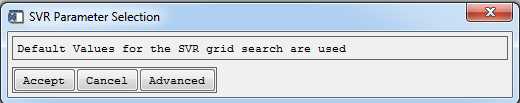
Default values for the grid search will be used to find ideal parameter values for the Gaussian kernel g, regularization C and loss function ε during model parameterization for all target categories. In most cases, default values already lead to high accuracies.
- Click Accept to proceed with SVR modeling using default values. The default values can be examined by clicking Advanced.
SVR modeling using advanced settings
The user may change the default values for the grid search or specify user defined parameters for the Gaussian kernel g, regularization C and loss function ε during model parameterization for all target categories.
- Click Advanced to continue with the advanced settings. The SVC parameterization dialog is now expanded.
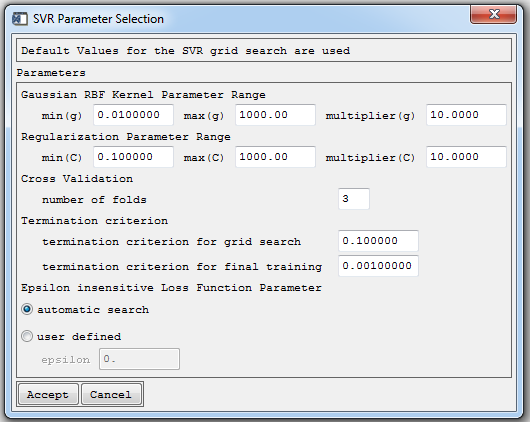
The user is now allowed to modify the grid search and may select:
min (g/C), max (g/C): Minimum and maximum values that define the range of the grid (g and C dimension).
Multiplier (g/C): Specifies the step size of the grid.
Cross Validation: The accuracy of results during the grid search is monitored by n-fold cross validation on the training data.
Termination criterion: Tolerance of termination criterion during grid search or final model training.
Epsilon loss insensitive Loss Function Parameter: Apply efficient search heuristic or enter user defined values.
- Adjust the settings if wanted and click Accept to proceed with SVR modeling using advanced settings.
synthMix-SVR will start with the SVR model training and model application for each target category by iterative processing. Corresponding SVR models and fraction maps will be saved in the output folder and opened in the EnMAP-Box Filelist.
The user may manually check the SVR parameters (e.g., selected parameter sets during grid search, training error, etc.) by opening the SVR models in the imageSVM Regression Application Menu.
- Applications > Regression > imageSVM Regression > View SVR Parameters For further information on SVR modeling using imageSVM, please refer to the imageSVM Regression Manual (van der Linden et al. 2014).
synthMix-SVR results
All generated files are saved in the specified output folder and opened in the EnMAP-Box:
- Original input spectral library, library labels and spectral image
- Synthetically mixed training data of the selected target categories
- SVR models of the selected target categories
- Single fraction map of each target category
- Stack of all fraction maps in a multilayer image
- (Optional) Stack of fraction maps with values between 0 and 100% in a multilayer image.
- (Optional) Stack of normalized fraction maps in a multilayer image. An output HTML-report will be opened in the standard browser for providing general information about the synthMix-SVR process. The user may display fraction maps in a RGB color composite and establish a link to the original spectral image
- Use the drag-and-drop functionality to display the test data set in the EnMAP-Box View Manager. The illustration below shows a composite of grass (R), tree (G) and impervious (B).
References
Burges, C.J.C. (1998). A tutorial on Support Vector Machines for pattern recognition. Data Mining and Knowledge Discovery, 2, 121-167.
Camps-Valls, G., Bruzzone, L., Rojo-Alvarez, J.L., & Melgani, F. (2006). Robust support vector regression for biophysical variable estimation from remotely sensed images. IEEE Geoscience and Remote Sensing Letters, 3, 339-343.
Chang, C.-C., & Lin, C.-J. (2011). LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology, 2.
Durbha, S.S., King, R.L., & Younan, N.H. (2007). Support vector machines regression for retrieval of leaf area index from multiangle imaging spectroradiometer. Remote Sensing of Environment, 107, 348-361.
Foody, G.M., & Mathur, A. (2004). A relative evaluation of multiclass image classification by support vector machines. Ieee Transactions on Geoscience and Remote Sensing, 42, 1335-1343.
Melgani, F., & Bruzzone, L. (2004). Classification of hyperspectral remote sensing images with support vector machines. IEEE Transactions on Geoscience and Remote Sensing, 42, 1778-1790.
Okin, G.S., Roberts, D.A., Murray, B., & Okin, W.J. (2001). Practical limits on hyperspectral vegetation discrimination in arid and semiarid environments. Remote Sensing of Environment, 77, 212-225.
Okujeni, A., van der Linden, S., Jakimow, B., Rabe, A., Verrelst, J., & Hostert, P. (2014). A Comparison of Advanced Regression Algorithms for Quantifying Urban Land Cover. Remote Sensing, 6, 6324-6346.
Okujeni, A., van der Linden, S., Tits, L., Somers, B., & Hostert, P. (2013). Support vector regression and synthetically mixed training data for quantifying urban land cover. Remote Sensing of Environment, 137, 184-197.
Rabe, A., Jakimow, B., van der Linden, S., Okujeni, A., Suess, S., Leitao, P.J., & Hostert, P. (2013). Simplifying Support Vector Regression Parameterisation by Heuristic Search for Optimal e-Loss. 8th EARSeL Workshop of Special Interest Group in Imaging Spectroscopy. Nantes, France.
Schölkopf, B., & Smola, A.J. (2002). Learning with Kernels - Support Vector Machines, Regularization, Optimization, and Beyond. Cambridge, Massachusetts: MIT Press.
Smola, A.J., & Schölkopf, B. (2004). A tutorial on support vector regression. Statistics and Computing, 14, 199-222.
Tuia, D., Verrelst, J., Alonso, L., Perez-Cruz, F., & Camps-Valls, G. (2011). Multioutput support vector regression for remote sensing biophysical parameter estimation. IEEE Geoscience and Remote Sensing Letters, 8, 804-808.
van der Linden, S., Rabe, A., Held, A., Wirth, F., Suess, S., Okujeni, A., & Hostert, P. (2014). imageSVM Regression, Manual for Application: imageSVM version 3.0. Humboldt-Universität zu Berlin, Germany.
License agreements
Redistribution and use of synthMix-SVR in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
1. Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
2. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
3. Neither name of copyright holders nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission.
THE SOFTWARE "synthMix-SVR" IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE REGENTS OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
imageSVM requires LIBSVM by Chih-Chung Chang and Chih-Jen Lin. LIBSVM is available at http://www.csie.ntu.edu.tw/~cjlin/libsvm. [Please note the separate copyright statement for LIBSVM]
Updated