Wiki
Clone wikienmap-box-idl / EnMAP-Box Manual
Overview
The EnMAP-Box is a license-free and platform-independent software interface designed to process hyperspectral remote sensing data, and particularly developed to handle data from the EnMAP sensor. Main features include:
- easy-to-use GUI, with drop-down menus, expandable tree-based file explorer and drag-and-drop capabilities.
- visualization and processing of hyperspectral image data and field/laboratory spectra.
- import and export from and to different data formats.
- in-built applications aimed at the processing of hyperspectral data, such as Support Vector Machines and Random Forests for classification or regression of image data.
- externally developed applications for EnMAP data processing, e.g. partial least squares regression or the calculation of different agricultural spectral indices. See Applications for an overview of all applications.
- a rich application programming interface (hubAPI) that allows to develop new EnMAP-Box applications easily.
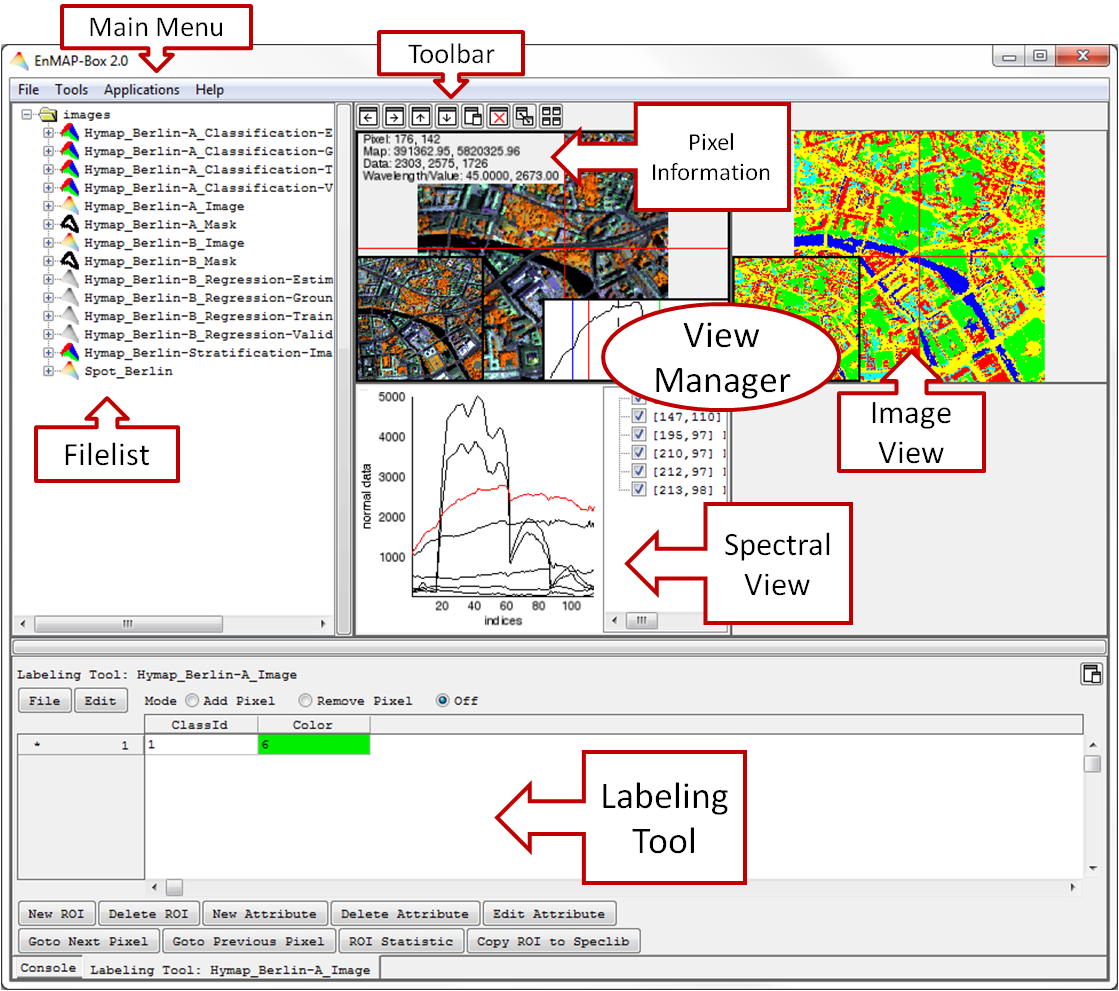
Graphical User Interface (GUI)
- The Main Menu offers several tools and applications to visualize, process and analyze spectral imagery or any image data.
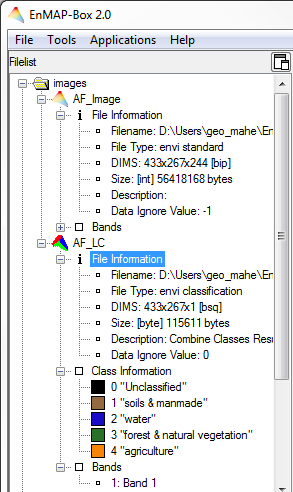
- The tree-based Filelist allows easy data organization. Every opened image or spectral data file includes its header information (File Information).
- A View Manager allows intuitive data visualization in several Image and Spectral Views. They can easily be added, deleted and arranged according to user’s desires or be detached from the View Manager
- Within the Image View, the Profile View, the Curser Information and an Overview Screen can be displayed or hidden.
- In the Spectral View the visualization and handling of spectral libraries is performed. The Image Views allow for an interaction with one or more spectral views.
File Management
Open Files
For image data and spectral data,
- select File > Open Image/SpecLib from the main menu and select your specific file. Furthermore, it is possible to get familiar with the EnMAP-Box by using the included test image data and spectral libraries,
- select File > Open > EnMAP-Box Test Images/SpecLibs from the main menu.
Context Menu
The context menu of the file list can be opened by right click on a file and allows you to
- Close single or all files
- Show Quick Statistics of the image or spectral library
- Edit Header Files of image data or spectral libraries
- Copy Meta Information of another file into the header of the current file
- Show the containing Folder
- Copy or Move the file to a defined destination
- Delete the file permanently
- Show the File Sources of the opened files
File Structure
Images
| Image Data | File Information | Bands |
|---|---|---|
 Spectral image Spectral image  Single band image Single band image  Mask Mask  Classification image Classification image | - filename with path - file type - dimensions of the file (DIMS) and interleave - size and data type - description* - data ignore value* - class information with color, value and name (classification images only) | all bands listed with their band numbers and wavelengths* |
* Note that, depending on the data, not all information will be available in the File List.
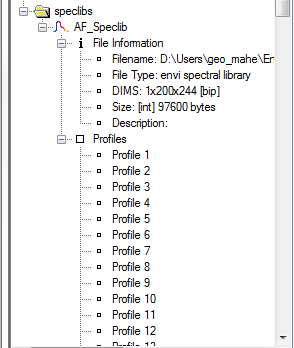
Spectral Libraries
The file tree also lists currently opened spectral libraries, with file information and the profiles with their names.
Other Files
All files which are not images or spectral libraries are grouped in folders according to their file extension. This allows keeping track e.g. of the model files that were created in the current session.
Data Types
Image data (i.e. spectral imagery, classification or regression images, reference areas and masks) are expected in binary format with an associated ENVI-style header file.
ENVI
description = {
Registration Result. Method: 1st degree...
12:34:26 2012]}
samples = 433
lines = 267
bands = 244
header offset = 0
file type = ENVI Standard
data type = 2
interleave = bsq
data ignore value = -1
sensor type = Unknown
byte order = 0
map info = {UTM, 1.000, 1.000, 665645....
coordinate system string = {PROJCS[“UTM..
default bands = {65,151,45}
wavelength units = Micrometers
wavelength = {
0.424000, 0.429000, 0.435000, 0.440000,...
...
Example of an ENVI-style header file
The EnMAP-Box allows the visualization of spectral (continuous) as well as classification (discrete) imagery data.
For more information refer to:
EnMAP-Box Data Format Definition
Moreover, routines for the import of additional information exist. Image data can be imported from TIFF, ASCII or Shapefiles (see Section 4.1) and will be physically written to disk in the current EnMAP-Box format.
Spectral data is expected to be stored in ENVI spectral library format. Applications in the EnMAP-Box requiring image data as input also allow spectral data. These are interpreted as single column pseudo images with
number of samples = 1
number of lines = number of spectra
number of bands = number of wavebands.
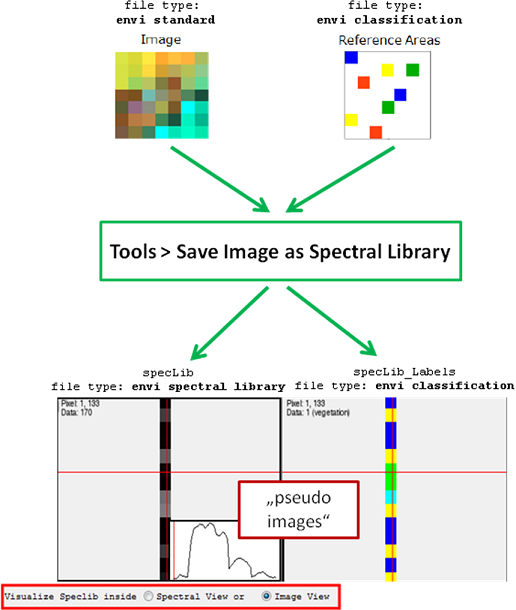
Furthermore, you now have the possibility to save and open EnMAP-Box spectral libraries. This allows you to store spectra from different sources and units in one data file. For the import of spectra, see Import of External Spectral Data.
Data Handling and Visualization
View Manager
Image data is visualized and handled in Image Views. Spectral data is visualized and handled in Spectral Views. Note that spectral data are interpreted as pseudo image and can be visualized in an image view as well. Both data types can be displayed by drag-and-drop of the file onto a free or an already occupied View in the View Manager.
- To open a new empty View, choose from one of these buttons



 in the Toolbar.
in the Toolbar.
The new View will be added to the defined location (e.g. left from the currently selected view). The views can be arranged using .
.
- To detach a view from the view manager or to move it to a new location, right click on the view and select Show View Menus (shortcut [m]). Use
 to move
to move
 to detach. To re-attach, simply close the window.
to detach. To re-attach, simply close the window.
 to remove the view.
to remove the view.
- To clear an Image or Spectral View, right click > Clear View.
- To open a new Spectral View, right click > New Spectral Library.
Image View
Navigation
For moving around in the image,
- hold the middle mouse button and move;
- left mouse click into the Overview Screen [o];
- use the arrow keys. For centering the image
- around the cursor double left mouse click;
- or use Shift + Return to move to scene center. For navigating to the scene edges,
- use Shift + Left/Right/Up/Down (arrow keys). For zooming the image,
- use the mouse wheel;
- use + and – on your keyboard;
- right-click (context menu) and choose Zoom to
- Pixel Size [1] : each image pixel as one pixel on the screen;
- Szene Size [2]: shows the full extent of the image;
- Szene Width [3]: width of the image is fully displayed;
- Szene Height [4]: height of the image is fully displayed;
- Level: determine a zoom factor.
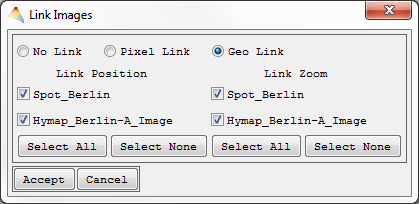
For linking image frame positions and zoom factors,
- select
 from the Toolbar and decide for
from the Toolbar and decide for
- Pixel Link (upper left pixel of one image is linked to upper left pixel of the other image) or
- Geo Link (use geocoordinates).
View Pixel Information
- Shortcuts: [i] and [p]
- left mouse click to select a pixel.
Information on the selected pixel is available as Cursor Information on top of the Image View. This includes the pixel’s image position, geographic coordinate (when available) and the value of the displayed band(s). - For activating/deactivating cursor information, open the context menu and choose Show/Hide Views > Show/Hide Cursor Information [i].

- Furthermore, the spectrum of a selected pixel can be visualized within a Profile View [p]. The current pixel profile can be
- linked to any open spectral view by using right click > Link Image Profile to Spectral View and
- copied to the linked spectral view by using right click > Copy Image Profile to linked Spectral View.
Band Selection
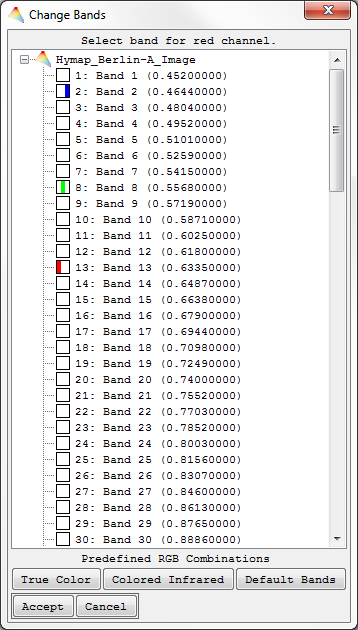
When opening multiple-band image files a Band Selection dialog appears and the user can manually select three spectral bands to be displayed in the respective RGB Combination, or choose between three options of predefined band combinations: True Color, Colored Infrared or Default Bands (requires respective header information). After accepting, the corresponding RGB/grey scale image will be displayed. By using the context menu (right click) in an open Image View, you can select Change RGB Bands to change the current band selection for the image.
Color Stretch
- Shortcuts:
- Ctrl+Mousewheel (stretch is based on full extent)
- Ctrl+Shift+Mousewheel (stretch based on view extent)
After opening the context menu in the image view (right click),
- click Set Data Stretch on > Full Extent or View Extent.
Spectral View
When you drop a Spectral Library onto an Empty View, you can either visualize it in a Spectral View or in an Image View (as pseudo image).
When you open the profiles in an Image View, they are interpreted as pixels arranged in a line.
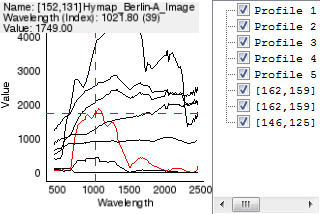
In a Spectral View, the profiles are plotted and listed in the Profile List.
- Hover over a profile to receive information on its name, wavelength (if available) and respective value.
- You can select profiles in the View and in the Profile List using
- the left mouse button;
- Ctrl (select single profiles);
- Shift (select several profiles).
- Right mouse button > Select all (Shortcut [Ctrl + A]).
- A selection allows you to
- Invert the current selection (right click > Invert Selection);
- Delete the profiles from the Profile List (right click > Manage Selection > Delete Selected Profiles);
- Show and Hide the selected Profiles (right click > Manage Selection > Show/Hide Selected Profiles).
- Copy profiles to another spectral view (right click > Manage Selection > Copy to Spectral View).
- Add profiles to a SOI (spectra of interest) when you have an open labeling tool (see Labeling Tool for Spectra).
In order to save the current spectral library in the EnMAP-Box Speclib data format
- right click > Save EnMAP-Box Speclib.
This represents a storage option for profiles from different sources and units. Another possibility for storage is the export to an ENVI spectral library by using
- right click > Export Profiles to ENVI Speclib.
Labeling Tool
For Images
In order to create regions of interest (ROIs) in an Image View, use
- right click > Show/Hide Views > Show Labeling Tool. This will open a tab with a labeling tool for the respective image.
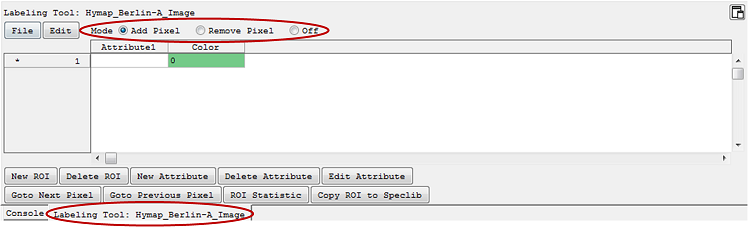
The regions are represented by the rows; each attribute belonging to a region is aligned in colums.
Labeling
- For labeling pixels/adding pixels to a ROI, select the Mode Add Pixel and choose a region on the left side (marked with *). In the respective Image View you can now select pixels of interest. By holding the Shift key you can quickly remove pixels. The number of pixels in the ROI are given in the column Color, where you can also change their color.
- The Mode Remove Pixel allows you to remove pixels from the ROI. By holding the Shift key you can quickly add pixels.
- The Mode Off deactivates the Labeling Tool.
Attributes
- For adding data in the attribute column, click on the empty space, type in the data and hit Return.
- The Button New Attribute lets you add attributes to the region. You can choose the name and data type (String, Integer or Decimal Number).
- Edit Attribute allows a later changing of name and data type.
- Delete Attribute removes attributes from the region.
- Edit on the upper left side allows calculations with the attributes.
Regions
- New Region adds a new ROI.
- Delete Region removes the currently selected ROI.
- Goto Next Pixel and Goto Previous Pixel lets you navigate through the selected ROI.
- ROI Statistics opens a HTML report with image statistics in the ROI.
- Copy ROI to Speclib allows you to fill a spectral library with the ROI’s spectra. For that, you have to open a Spectral View at first.
File I/O
- Use File > Open ROIs / Save ROIs to handle the data in the enmapROI format.
- Use File > Import ROIs from Label Image / Shapefile to receive ROI information from other sources.
- Use File > Export ROIs to
- Label Image
- Classification Image
- Shapefile
For Spectra
In order to create spectra of interest (SOIs) in a Spectral View, use
- right click > Show Labeling Tool. This will open a tab with a labeling tool similar to that for ROIs.
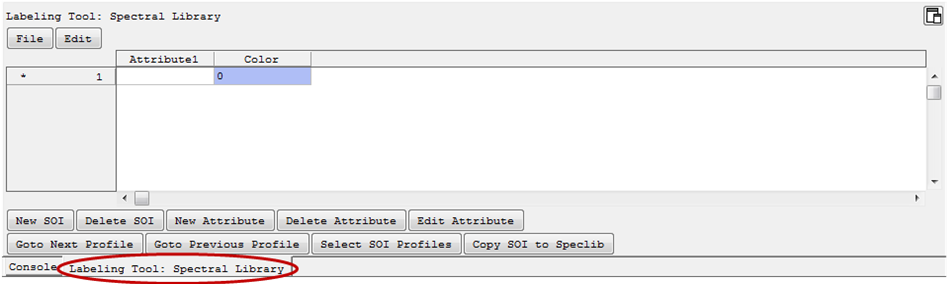
Labeling
For labeling spectra/adding spectra to a SOI (spectra of interest), select a SOI on the left side (marked with *). In the respective Spectral View you can now select spectra of interest and add them to the selected SOI by
- right click > Manage Selection > Add to current SOI.
- use right click > Manage Selection > Remove to current SOI to remove selected spectra from a SOI
- The number of spectra in the SOI are given in the column Color, where you can also change their color.
File I/O
- Use File > Open SOIs / Save SOIs to handle the data in the enmapSOI format.
- Use File > Import SOIs from Label Image (pseudo image) to import a labeled pseudo image.
- File > Export SOIs to Label Image (pseudo image) allows you to create a regression dataset and/or save several attributes of the SOIs saved as a pseudo image.
- File > Export SOIs to Classification Image (pseudo image) creates a pseudo image with classification labels.
Interaction with spectral views
- Copy SOI to Speclib allows you to fill another spectral library with the SOI’s spectra.
- Select SOI Profiles will create a selection in the spectral view.
Import Data
Import Image Data
External image data in TIFF or ASCII format and Shapefiles can be imported with the EnMAP-Box. The corresponding image files are transformed into the current EnMAP-Box format and physically written to disk.
Import TIFF / GeoTIFF
- Select File > Import > Image > TIFF / GeoTIFF Image.
- Choose an Input TIFF File and specify the name and path of the output file in EnMAP-Box format. The imported file will appear in the Filelist.
Import ASCII Lists
For ASCII file types a set of header lines specify the dimensions of the image file to generate.
ASCII lists consist of two columns that specify the location in the image (x-y-pixel coordinates) and additional columns with attributes corresponding to these locations. If more than one additional column exists, e.g. when multiple field measurements were performed at one position, individual reference area files are saved for each variable. Testdata are available in the following directory:
EnMAP-Box/enmapProject/enmapBox/resource/testData/ascii/ASCII_list.txt
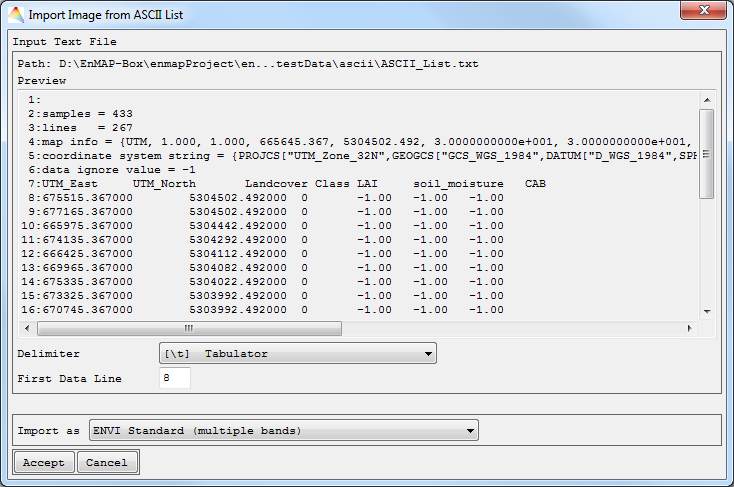
- Select File > Import > Image > from ASCII List.
- Choose an Input ASCII File and Accept.
- The data preview dialog will show you the first lines of your data. Now specify the Delimiter of the columns, define where the First Data Line begins and choose whether to Import the image as ENVI standard, ENVI classification or EnMAP-Box Regression. Click Accept.
- In the new dialog, under “Columns”, you can choose one (single band) or more (multiple bands) band value columns to be represented in the image.
- Two Coordinate Columns have to be defined, and the Coordinate Type with Geo- or Pixel Coordinates.
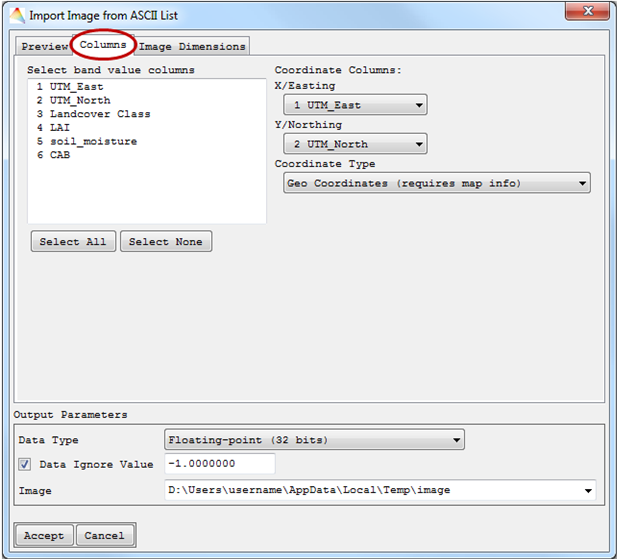
- Finally, the Image Dimension has to be specified. Either it will be a Copy from Reference Image or a New definition. When the ASCII file already contains information on geo coordinates, they will be used for the image. Otherwise Without Georeference / Map Info will be automatically selected.
- Output Parameters include Data Type, Date Ignore Value and path of the Image.
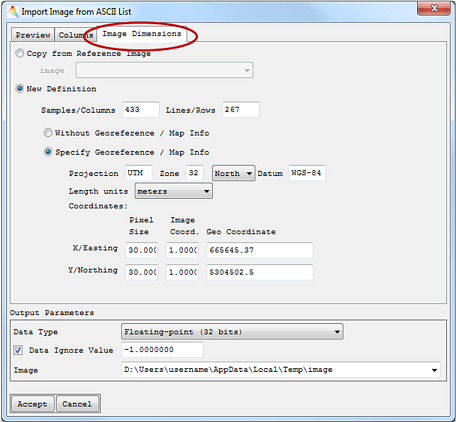
The imported file will appear in the Filelist.
Import ASCII Grids
ASCII Grid files have the relevant pixel information arranged in a grid.
When importing ASCII raster files/ASCII Grids, the resulting image will be of the same dimension as the raster file. Testdata are available in the following directory:
EnMAP-Box/enmapProject/enmapBox/resource/testData/ascii/ASCII-GRID_AF_LAI.txt
- Select File > Import > Image > from ASCII Grid.
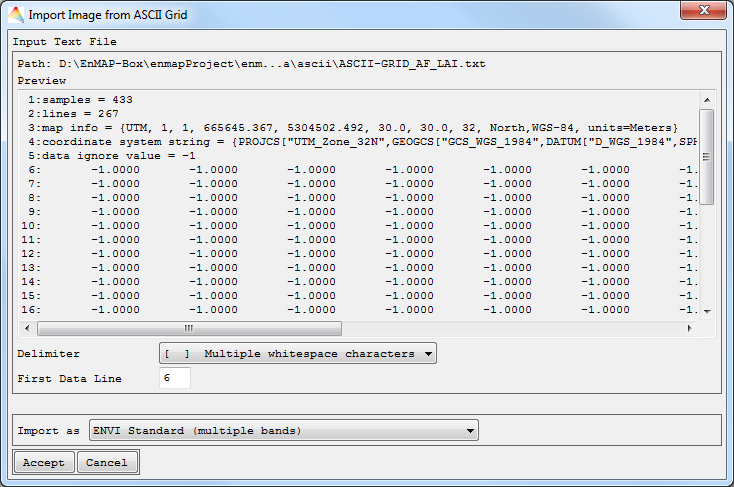
- Choose an Input ASCII File and Accept.
- The first lines of the data are shown. You have to choose Delimiter, First Data Line and whether to Import the image as ENVI standard, ENVI classification or EnMAP-Box Regression. Click Accept.
- Finally the Image Dimension has to be specified. Either it will be a Copy from Reference Image or a New Definition. In your New Definition the number of Bands has to be specified. When the ASCII file already contains information on geo coordinates, they will be used for the image. Otherwise Without Georeference / Map Info will be automatically selected.
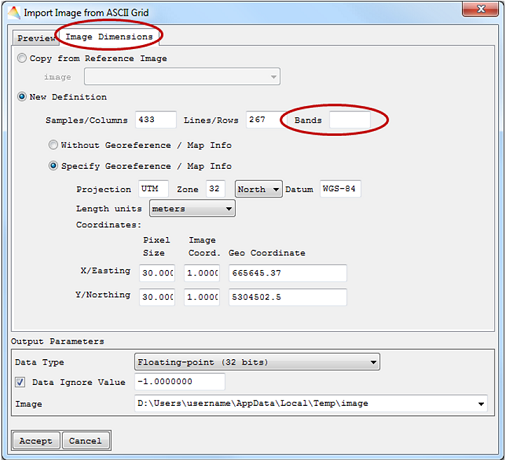
The imported file will appear in the Filelist.
Import of External Spectral Data
The EnMAP-Box offers the opportunity to import external spectral data in ASCII format.
- Select File > Import > Spectral Library
- from ASD Field Spec Files;
- from ASCII List (ENVI Format);
- from CSV Table.
- ASD Field Spec Files
- Choose the ASD binary files to be imported, and define where to save the Output spectral library.
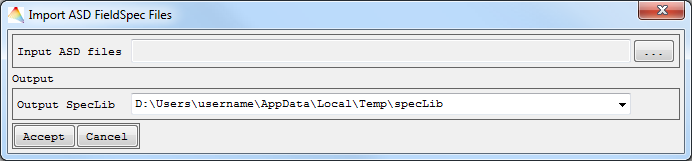
The file will appear in the Spectrum List.
ASCII List (ENVI Format)
Testdata are available in the following directory:
EnMAP-Box/enmapProject/enmapBox/resource/testData/ascii/ESL_Ascii.txt
- Choose the Input ASCII File and define where to save the Output spectral library.
The file will appear in the Filelist.
CSV Table
Testdata are available in the following directory:
EnMAP-Box/enmapProject/enmapBox/resource/testData/ascii/SL_ASCII.txt
- Choose the Input ASCII File.
- The data preview dialog will show you the first lines of your data. Now specify the Delimiter of the columns and where the First Data Line begins.
- If your data contains a Wavelength Column, check the respective button and define column number and the Wavelength Units.
- Select the Data Type and the path where to save your SpecLib.
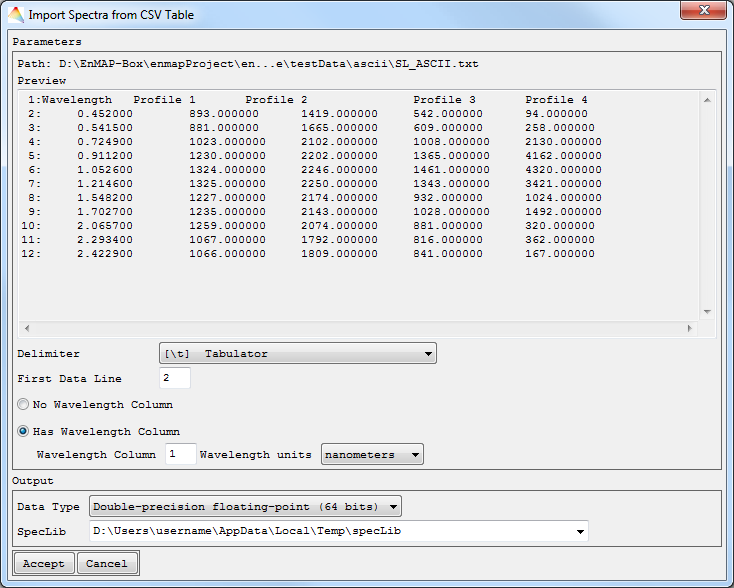
The file will appear in the Filelist.
Export Image to TIFF/GeoTIFF
- Select File > Export > Image to TIFF/GeoTIFF.
- Choose an Input Image and where to save the Output TIFF.
The file will be saved to disk.
Tools
Image Statistics
- Select Tools > Image Statistics.
- Choose the Image you want to investigate.
- Optional: Apply a Mask , choose a Stratification image or include histogram plots in the report.
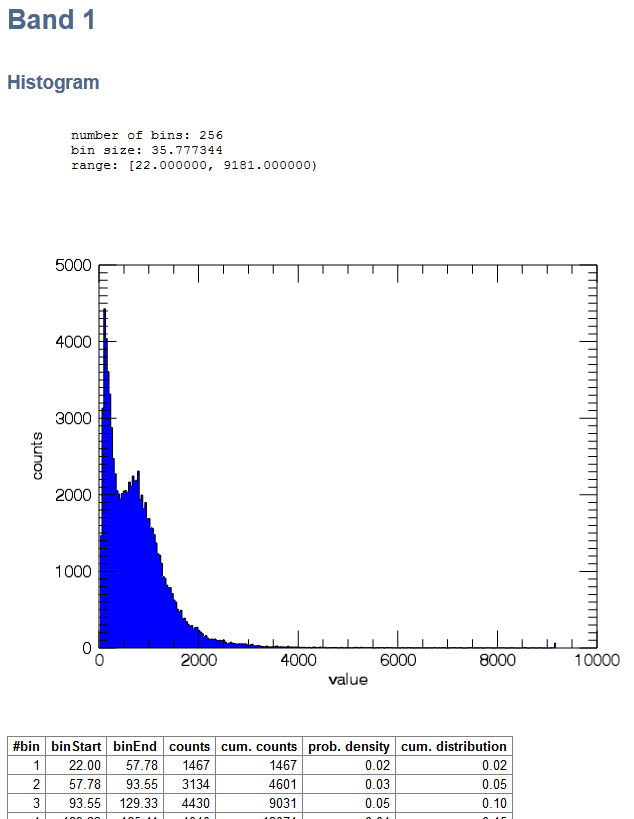
The Image Statistics will be calculated and appear in your HTML Browser. Note that, depending on the input image, this may take a while. Listed are the File Information, an Overview with band statistics and the statistics for each band. In a table the start and end of each bin is listed, together with the absolute and relative frequencies of the pixels. When you chose a Stratification image, the statistics are presented in this order:
- 1. Overview
- Band values (statistics for each band)
- ROI: Class 1 (band-wise statistics for ROI 1)
- ROI: Class 2 (band-wise statistics for ROI 2, etc.)
- 2. Band 1
- Histogram table for band 1
- Regions of interest
- Class 1 (histogram table for ROI Class 1 at band 1)
- Class 2 (histogram table for ROI Class 2 at band 1)
- 3. Band 2 …
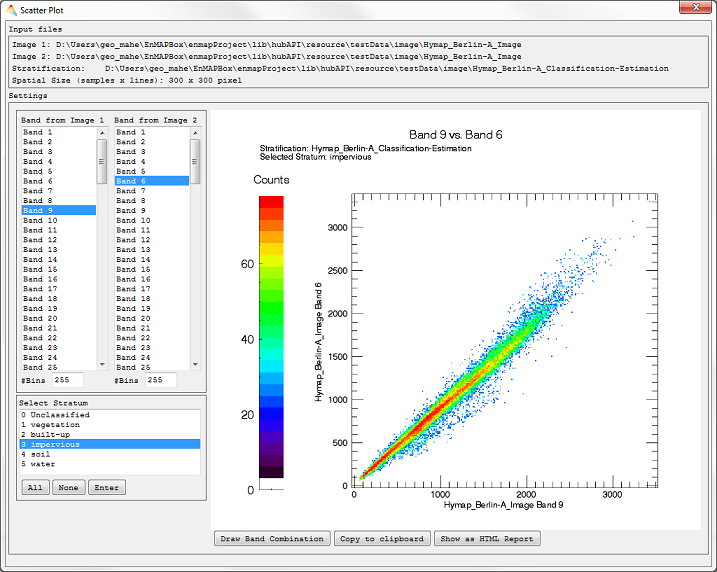
Scatter Plot
- Select Tools > Scatter Plot.
- Input: Two Images have to be defined, either the same image, in case you want to plot different bands of the same image, or two different images.
- Optional: Apply a Mask or choose a Stratification Image (to plot pixels of different strata).

- Accept, and a Scatter Plot dialog opens listing your previous specifications above.
- Choose which bands to plot and click Draw Band Combination. If previously defined, Select Stratum.
- Click Show as HTML Report to open the plot in your HTML browser or Copy to clipboard to copy the plot.
Scale Image Data
Input to Output Range
- Select Tools > Scale Data > Input to Output Range
- Choose an image to be scaled.
- Optional: Apply a Mask.
- Decide whether to define the Input Range either
- by Data Values (minimum and maximum of your input data defined manually) or
- by Percentiles and define the corresponding range. By default, the percentiles will be calculated for each band individually (otherwise click Advanced).
- Define the Output Range to which your Input shall be scaled, and decide whether to Trim Outliers to Min/Max (check) or not (uncheck). If this is checked, the pixel values beyond the Input Range will not be linearly scaled and are fixed to the minimum resp. maximum value.
- Define the Data Ignore Value and the name of the Scaled Image.
- Click Advanced to calculate the percentiles from full image, bandwise or from an external image. Here you can also define the Data Type of the output image.
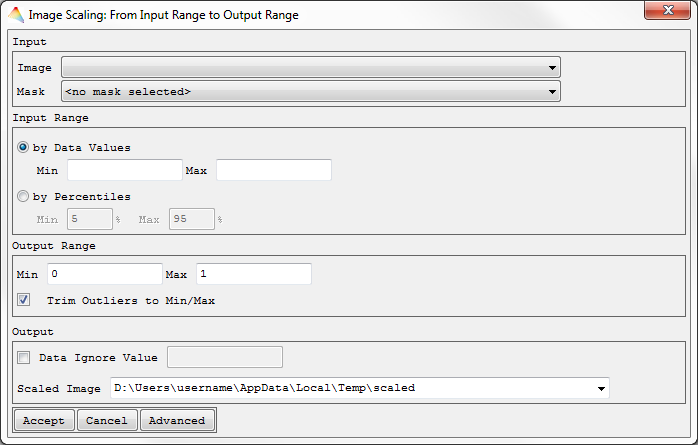
The image will be scaled and appear in the Filelist.
Z-Transformation
- Select Tools > Scale Data > Z-Transformation.
- Select the Image to be scaled.
- Optional: Apply a Mask.
- Choose one of the options to get the parameters mean and standard deviation.
- Select Use Mean and Standard Deviation from Image Statistics, by default both parameters will be calculated bandwise (otherwise click Advanced).
- Select Set Mean and Standard Deviation manually. E.g., when you define the mean of the input image to be 0 and the standard deviation to 1, the image is already z-standardized and will not be changed.
- Define the Output Data Type and the name of the Scaled Image.
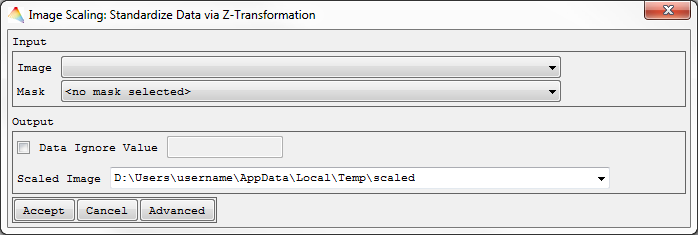
The image will be scaled and appear in the Filelist.
Random Sampling
- Select Tools > Random Sampling.
- Choose the Image of interest.
- Optional: Apply a Mask or choose a Stratification image (generate sampling points from selected strata).
- By selecting Advanced, you can specify
- how the Random Sequence Seed will be created, it will either be a unique sample (Current System Time) or a reproducible sample (Constant), and
- how pixels with data ignore values are excluded: choose ALL bands to exclude pixels having the data ignore value set in each band, or choose ANY band to exclude pixels having the data ignore value set in at least one band.
- Accept, and in the next dialog
- choose a defined sample size in pixels (Absolute Sampling) or a percentage of all pixels (Relative Sampling).
- In case you defined a Stratification image, equalized and proportional sampling will yield the same amount of pixels for each stratum. Otherwise you can select Disproportional Sampling.
- Define where to save the Random Sample and optionally to save the sample Complement.
Non-stratified sampling
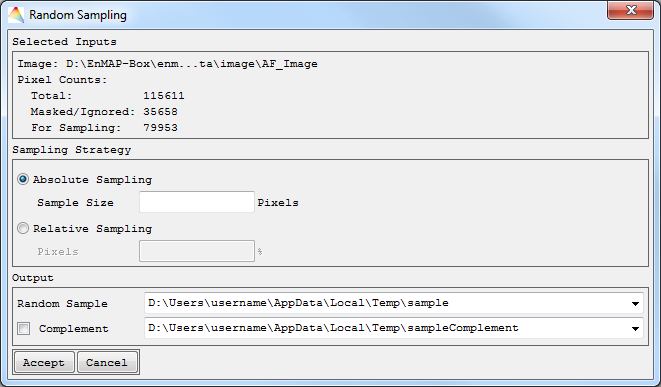
Stratified sampling
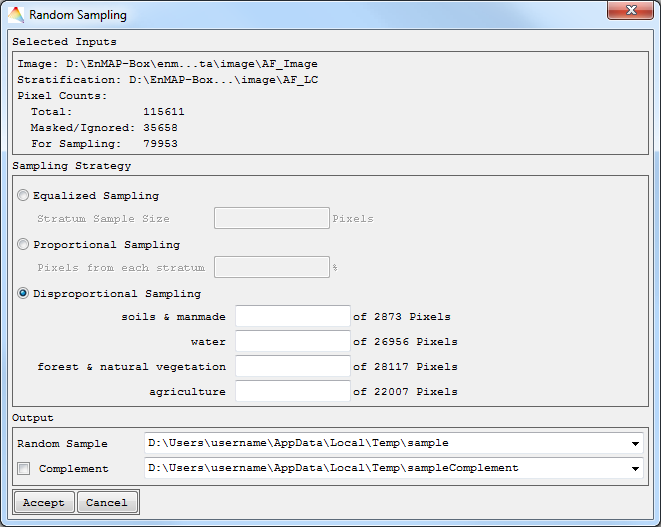
Your random sample will appear in the Filelist.
Spatial / Spectral Subset
- Select Tools > Spatial/Spectral Subset.
- Choose the Image of interest.
- Optional: Apply a Mask.
The dialog ImageSubsetting opens with the Input Info on top.
- For Spatial Subsetting, choose Pixel- or Geo Coordinates of your subset, click the Preview button and a rectangle will indicate the subset.
- For Spectral Subsetting, choose Spectral Subset and select one or more bands for your subset (use Ctrl and Shift).
- Define where the Output Image is to be saved.
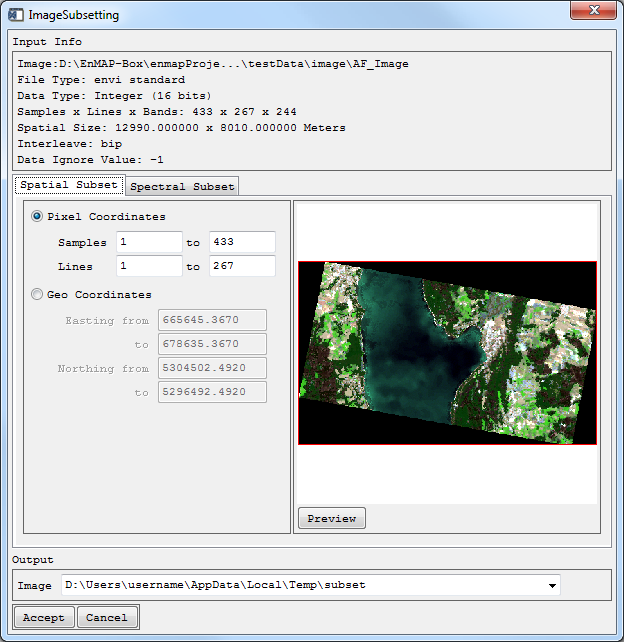
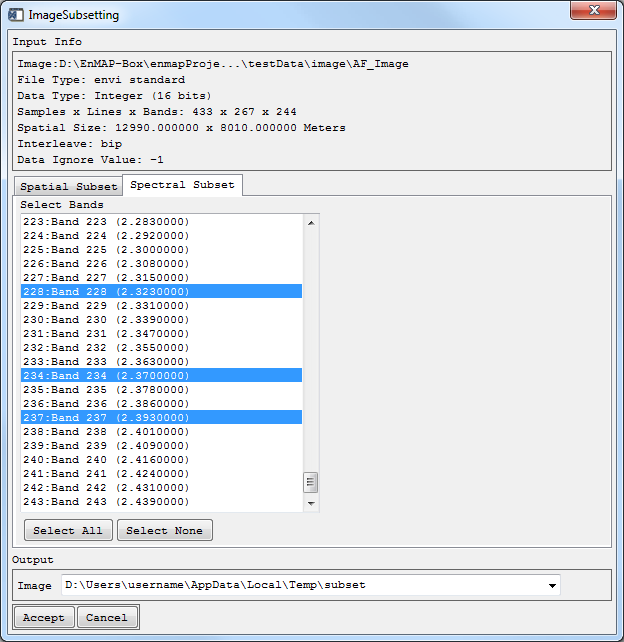
Your subset will appear in the Filelist.
Spectral Resampling
- Select Tools > Spectral Resampling.
- Choose the Image or Speclib to be resampled. In this example, the input is a Landsat 5 speclib and it is supposed to be resampled to Landsat 8 OLI.
- For the resampling, spectral response functions of a sensor are needed. Either choose an existing response functions file (Spectral Response Function), or choose the second option where the response functions will be calculated from the FWHM information in the header of the file (Band Wavelength + FWHM).
- Now choose where to save the Resampled Image/Speclib.
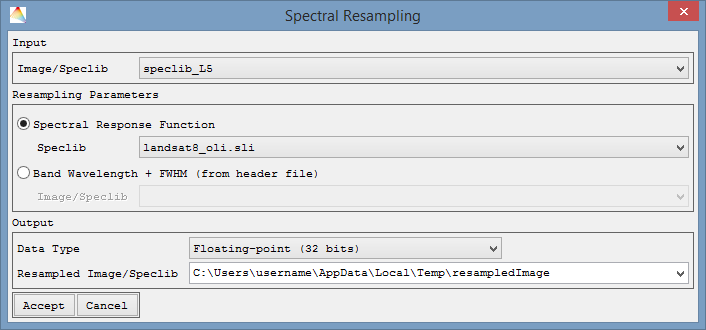
- Next you can (1) select which of the output bands you want to use and (2) create Preview plots, to get an idea on the quality of the resampling.
- If your input image/speclib did not have FHWM information in the header file, which are however needed to create the preview plots, you can here select a response functions file, corresponding to the input.
- Here, too, you can subset the bands such that they correspond to the input spectra.
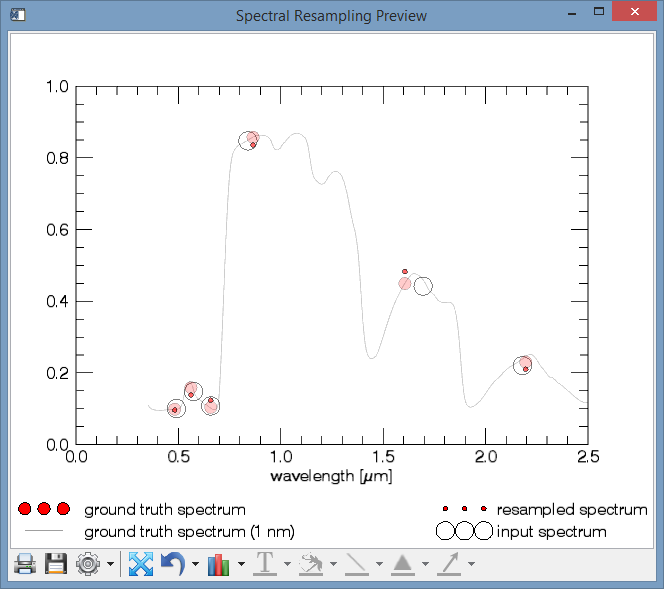
- Two plots will be created.
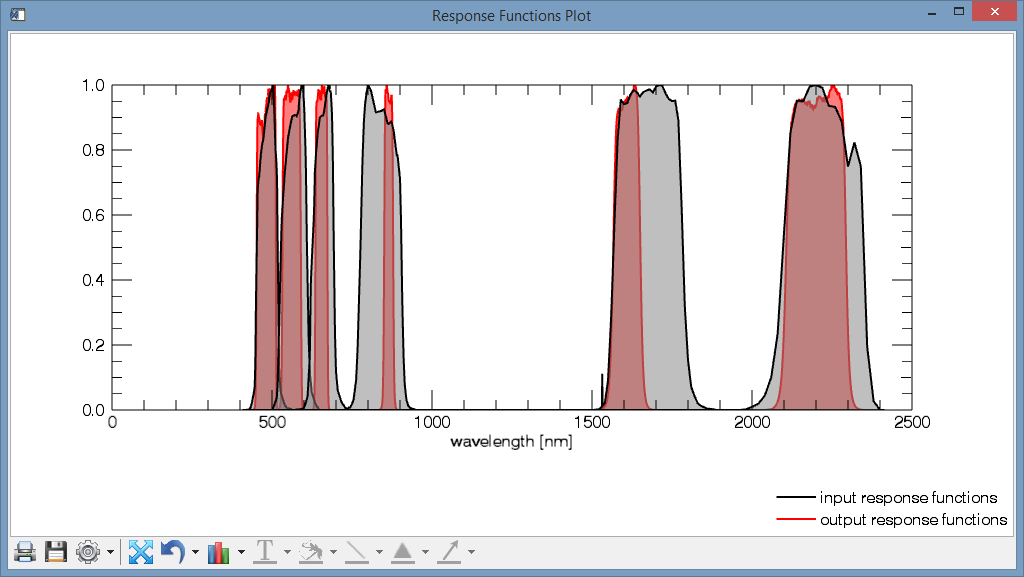
- A Spectral Resampling Preview Plot, depicting (1.) how the resampled spectrum (in this case L5 -> L8) would look like, given an input spectrum (2.). For comparison, the ground truth spectrum is plotted, i.e. how the output sensor would record the true surface (3.), and how the ground truth looks like in 1 nm resolution (4.).
- A Response Functions Plot, comparing the reponse functions of the input sensor with the response functions of the output sensor.
Reclassify Image
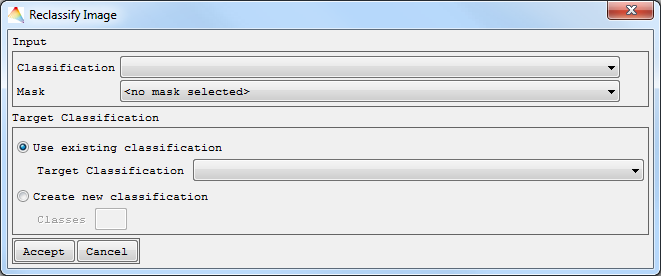
- Select Tools > Reclassify Image.
- Choose your Input Classification.
- Optional: Apply a Mask.
- Choose one of the possibilities for reclassification.
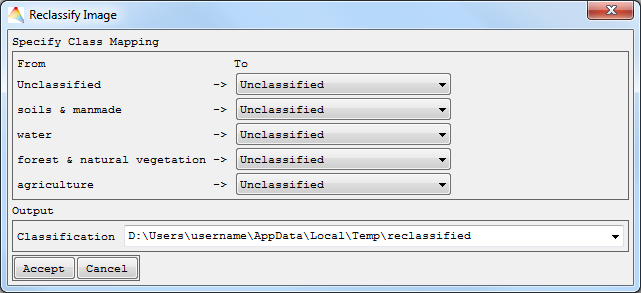
- Use existing classification as Target Classification, then Accept, specify the new memberships and define where to save the new one.
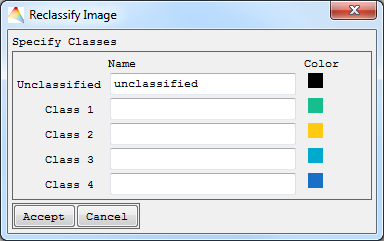
- Create new classification. Define the number of Classes, then Accept. Now specify the names of the new classes and the respective colors, then Accept. Finally select the new memberships and define where to save the new Classification.
Your new classification will appear in the Filelist.
Save Image as Spectral Library
- Select Tools > Save Image as Spectral Library.
- Choose your Input Image from which the spectra are to be used.
- Check Reference Areas if you want to generate the spectral library (SpecLib) from the Image using reference points.
- Optional: Apply a Mask.
- Define where to save the Output SpecLib and Accept.
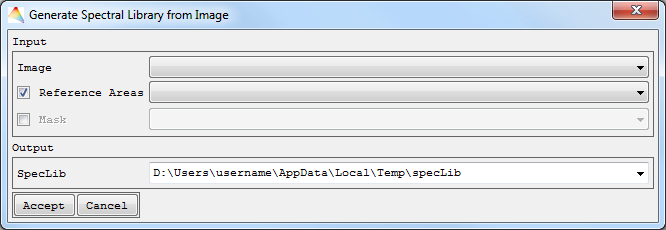
The labels of the spectra speclib_Labels will appear under the folder images, and the spectral library speclib appears under speclibs.
Apply Mask
- Select Tools > Apply Mask.
- Choose the Input Image and which Mask shall be applied to it, finally define where the Masked Image is to be saved and specify if the data ignore value in the created image is to be used from the input image or defined by a new value.
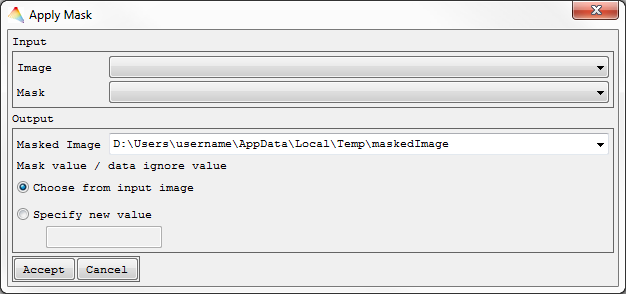
Your masked image will appear in the Filelist.
Stack Images
- Select Tools > Stack Images.
- Choose the images to stack.
- The Data Type of your stack has to be selected and optionally a Data Ignore Value defined. By default, the Data type of the image with the largest data type size will be used.
- Define where to save the Stack Image.
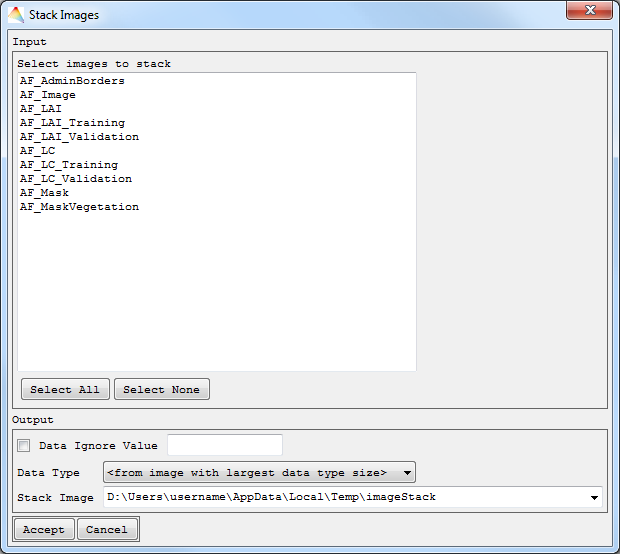
The image stack will appear in the Filelist.
Applications
Classification
imageRF
For Random Forests Classification refer to the imageRF Manual.
imageSVM
For Support Vector Classification refer to the imageSVM Classification Manual.
Regression
Least-Squares Regression
Parameterize
- Select Applications > Regression > Least-Squares Regression > Parameterize.
- Choose the Input Image and the Reference to be used.
- Optional: Apply a Mask.
- Select the Regression Model Type, either using Univariate or Multivariate Least Squares, and Accept.
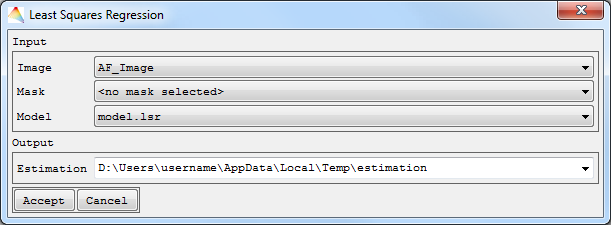
- A new dialog opens.
- If you used Univariate Least Squares, choose Polynomial Degree and Select Input Feature, and finally define the path to save the model file.
- If you used Multivariate Least Squares, Select Input Features will allow choosing several bands for a 1st degree polynomial. Define where to save the model file.
- Accept, and the model parameters will show up in your HTML browser.
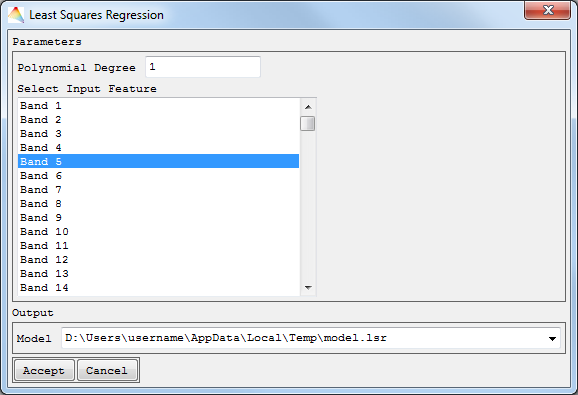
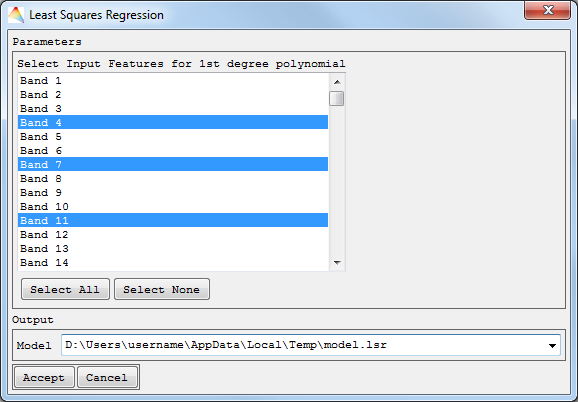
Univariate and Multivariate Least Squares Regression
Apply to Image
- Select Applications > Regression > Least-Squares Regression
Apply to Image.
- Choose the Input Image.
- Optional: Apply a Mask.
- Select the Model to be applied to the Image (by default the last Model is selected).
- Now define where to save your Estimation and Accept.
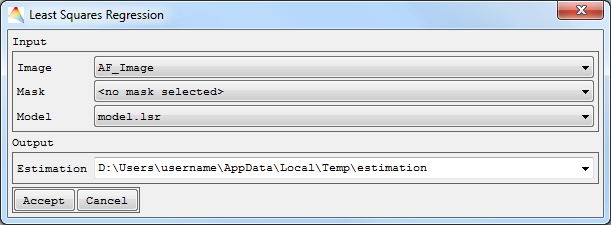
The file will appear in the Image List named estimation.
Fast Accuracy Assessment
- Select Applications > Regression > Least-Squares Regression > Fast Accuracy Assessment.
- Define the Model to be applied to the Image (by default the last Model is selected).
- Choose the Input Image and Reference Areas, and Accept.
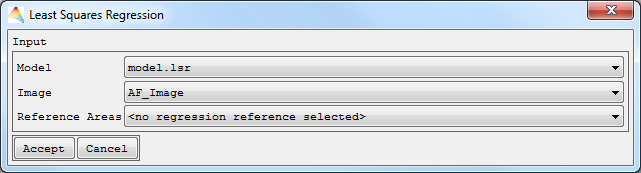
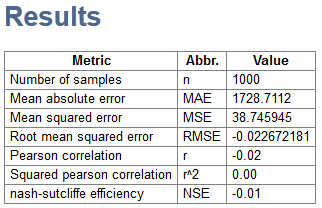
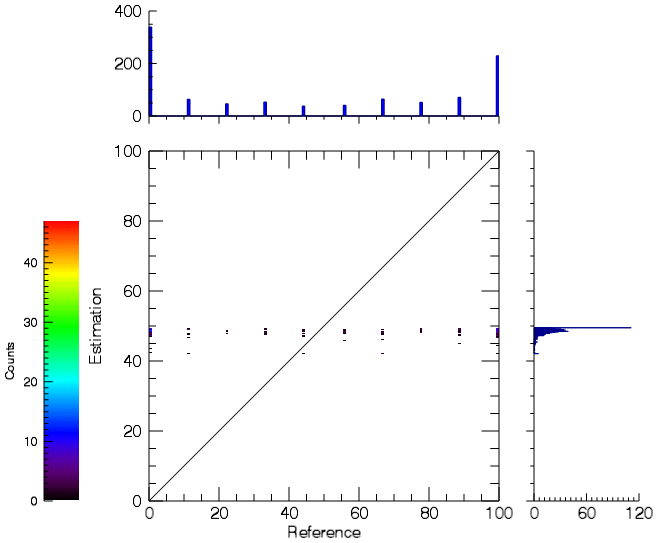
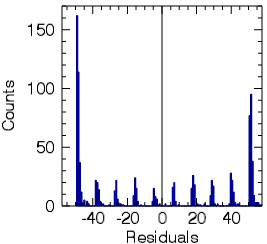
- A HTML report will open with information about accuracy measures such as RMSE and MAE, as well as the Scatterplot and Residual Histogram.
View Parameters
- Select Applications > Regression > Least-Squares Regression > View Parameters.
- Define the Model.
- A HTML report will open and show the Constant and Coefficients of the model.
imageRF
For Random Forests Regression please refer to the Manual or Application Tutorial.
imageSVM
For Support Vector Regression please refer to the Manual or Application Tutorial.
Accuracy Assessment
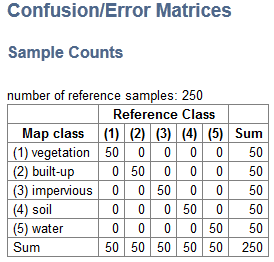
Classification
- Select Applications > Accuracy Assessment > Classification.
- Choose the Classification Estimation and Reference image.
- Optional: Apply a Mask or choose a Regions of Interest image (to get accuracy measures for each region).
- By clicking Advanced, you can decide to Save Errors.

- Accept, and an HTML report will open with information about several accuracy measures and error matrices. Formulars are taken from: Assessing the Accuracy of Remotely Sensed Data. Principles and Practices, Second Edition, Russell G. Congalton and Kass Green, 2007.
Regression
- Select Applications > Accuracy Assessment > Regression.
- Choose the Regression Estimation and a Reference image.
- Optional: Apply a Mask or choose a Stratification image (to get accuracy measures for each stratum). Click Advanced to change the Number of histogram bins, to Use equal bin ranges for all strata or Save Errors.
- A HTML report will open with information about accuracy measures such as RMSE and MAE, as well as the Scatterplot and Residual Histogram.
Digital Filter
Savitzky-Golay-Filter
The Savitzky-Golay smoothing filter (or digital smoothing polynomial) is a spectral data filtering method, which is suitable for smoothing data with a noisy signal.
- Select Applications > Digital Filter > Savitzky-Golay Filter.
- Choose the Input Image of interest and define the Filter Parameters (integers):
- filter width: An integer specifying the number of data points to include in the filter. Note that the filter width needs to be an odd number and larger than the polynomial degree, and that larger values produce a smoother result at the expense of flattening sharp peaks.
- polynomial degree: An integer specifying the degree of smoothing polynomial. Lower values will produce smoother results but may introduce bias, higher values will reduce the filter bias, but may overfit the data and give a noisier result. Note: degree must be less than the filter width.
- derivative order: An integer specifying the order of the derivative desired. For smoothing, use order 0. To find the smoothed first derivative of the signal, use order 1, for the second derivative, user order 2, etc. The order needs to be smaller or equal to polynomial degree.
- Define where to save the Result Image, Accept and the filtered image will appear in the Filelist.
imageMath
For information on the imageMath Calculator please refer to the respective Manual.
Updated