Wiki
Clone wikiACID - Artefact correction in diffusion MRI / Diffusion signal fitting models
Diffusion signal fitting models
The Tensor Fitting tool is an academic software to estimate the diffusion tensor. The methods available to estimate the tensor are: ordinary least squares, weighted least squares, and robust tensor fitting.
Fit Diffusion tensor pre-processing
- Use ECMOCO to correct for EC and motion artefacts.
- Define two variables in matlab, which cover the diffusion directions (3xN matrix) and b-values (1xN vector), before running the Fit Diffusion tensor toolbox. The “i-th” column (component) must correspond to the vector of the diffusion gradient (and the b-value) of the “i-th” image in the DTI dataset. If the b-value for the low-b-value images is unknown, type b=1, and if its diffusion gradient direction is unknown, type a random direction, which is normalised to 1.
Use Tensor Fitting
- dMRI images: Load pre-processed DTI images. The order of images can be random, as long as the diffusion directions and b-values are loaded in the same order.
- b-values: Load the b-values (1xNvector) in the same order as the DTI images.
- b-vectors: Load the b-vectors (3xN array) in the same order as the DTI images.
- Region of interest image (optional): This entry is optional. Note that if your had acquired HARDI data (and/or high-resolution data), it is recommened to provide a mask unless you believe that you have got enough memory!
- Fitting algorithm: Here you can choose between three tensor fitting algorithms:
- Ordinary least squares (OLS): This method will deliver the highest SNR but is most sensitive to outliers.
- Weighted least squares (WLS): This method will (to some degree) account for the Rician noise distribution.
- Robust fitting: This method will down-weight outliers and is most efficient for reducing the effect of outlier-related bias in dMRI data (see e.g. Mohammadi et al. (2013)).
- Non-Linear Least Squares (NLLS): XXXX
- Axial Symmetric (AX-DKI): XXXX
- Parallel programming: Note this option requires the parallel programming toolbox. Specify number of cores for parallel programming. No indicates that no parallel programming is used.
Defaults (Changeable in /config/local/acid_local_defaults.m):
- Write option: Write fitted diffusion-weighted images (DWI). This option provides DWIs with reduced noise, but keep in mind that the modeled DWIs are limited by the DTI model used.
- Weights option: Option to save the weights that were generated during robust fitting on a voxel-by-voxel basis as a 4d volume.
- Display option: Option to display weights during robust fitting. Might be instructive to look at it once in a while.
- Tikhonov regularization constant: This is the value of a Tikhonov regularization constant and should not exceed 1e-4 otherwise it can bias the tensor estimates. It is also related to the number of iteration taking place.
- Reorient matrix: A global matrix to reorient the diffusion vectors. This might be useful, if the diffusion gradients are defined in a different coordinate systems than the acquired images.
- Smooth the mask: This option reduces holes in the brain mask by smoothing the mask or the tissue probability maps. If it is set to zero no smoothing is used.
- Confidence interval: Confidence interval for robust fitting.
- Minimal diffusivity: Threshold for minimal diffusivity.
- Residual smoothing: This variable determines the smoothing that is applied on the residuals; smaller maxk results in more smoothing.
- Deviation of logarithm of the signal: Standard deviation of logarithm of the signal outside the brain. This measure is used if the noise cannot be estimated from outside the brain (see brain mask option).
- Write eigenvector option: Determines whether to write out all eigenvectors of the diffusion tensor, i.e. 1st, 2nd, and 3rd eigenvector and eigenvalues. The filename will be extended by the number of the eigenvector components and the eigenvector number, i.e. "filename-ij.img" with "i" being the eigenvector components and "j" being the eigenvector number.
- Number of fit iterations (only NLLS): Choose the number of fit iterations.
Background
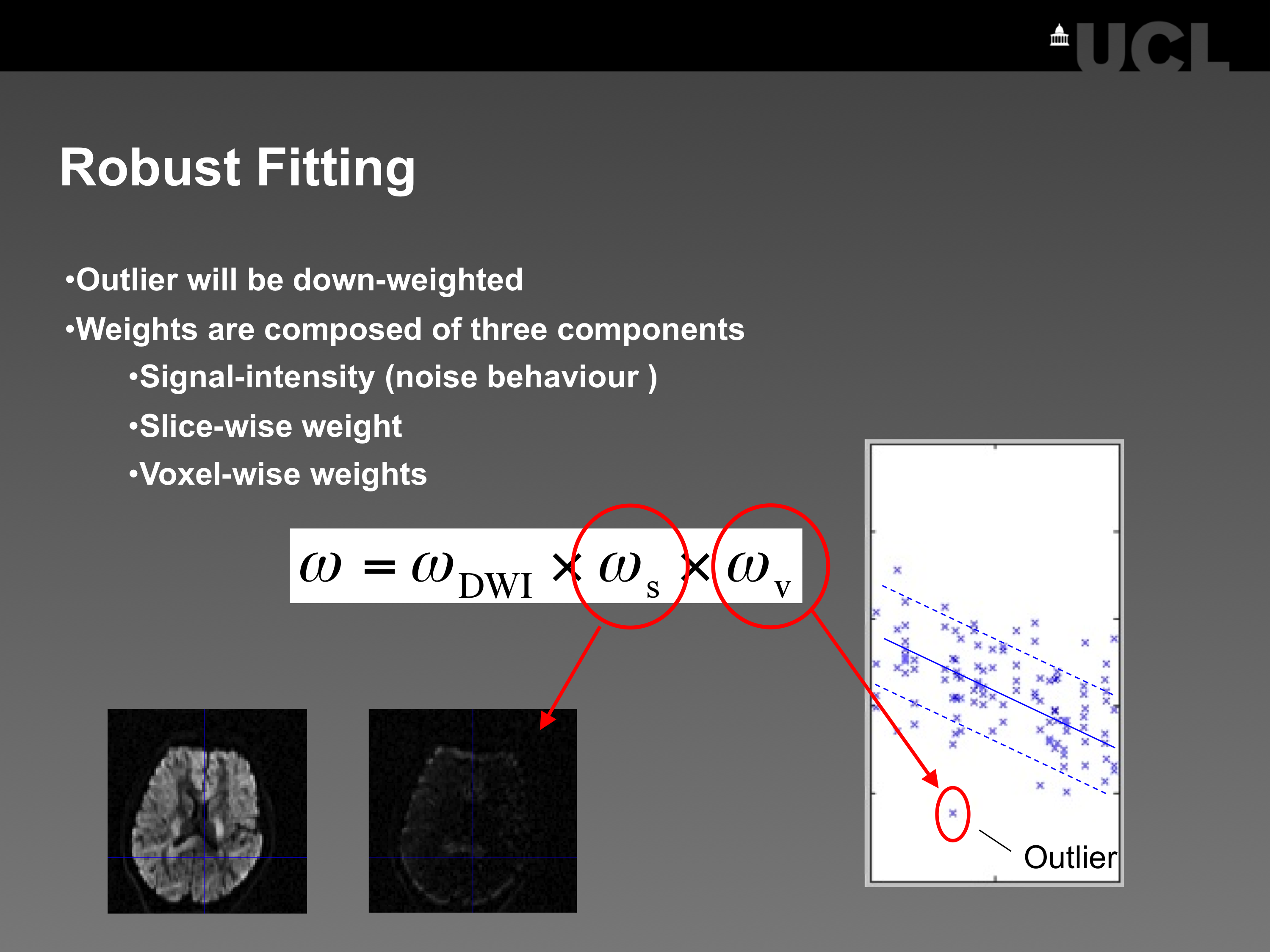
Fig. 1: The robust fitting model down-weights two types of outliers that fall far outside the expected normal distribution of residuals C (the so-called “confidence interval”): (a) Voxel-wise outliers are addressed with a term that changes from voxel-to-voxel (ωv). This measure is noisy; to reduce the noise the limits for the confidence interval C will be smoothed (see Defaults “smoothing the residuals”). (b) Signal-dropout within a whole slice (e.g. due to subject motion) is addressed with an additional term that only changes from slice-to-slice (ωs) and thus is less noisy.
The basic idea of the robust-fitting method is based on the paper of Zwiers (2010), where different types of weighting functions are combined using a Gaussian weighting scheme. Robust fitting is recommended if your data suffers from outliers, e.g. due to physiological noise (e.g. Mohammadi et al. (2013a)) or subject motion (e.g. Mohammadi et al. (2013)). It works well if your outliers are sparsely distributed along different diffusion directions (Zwiers (2010)) and you acquired enough data (N>30, where “N” is the number of images in the DTI sequence, Chang et al. (2005)). Note that if the amount of outliers is too high in a dMRI dataset, outlier-rejection might worsen the data quality instead of improving it. In our toolbox, we take care of this problem by checking the condition number of the diffusion-tensor design matrix, which down-weighting of individual data points. Note also that for data with low SNR (SNR < 4) the Gaussian noise distribution might be violated and the linear tensor fitting methods in this toolbox might not be valid anymore.
Example in spinal cord DTI
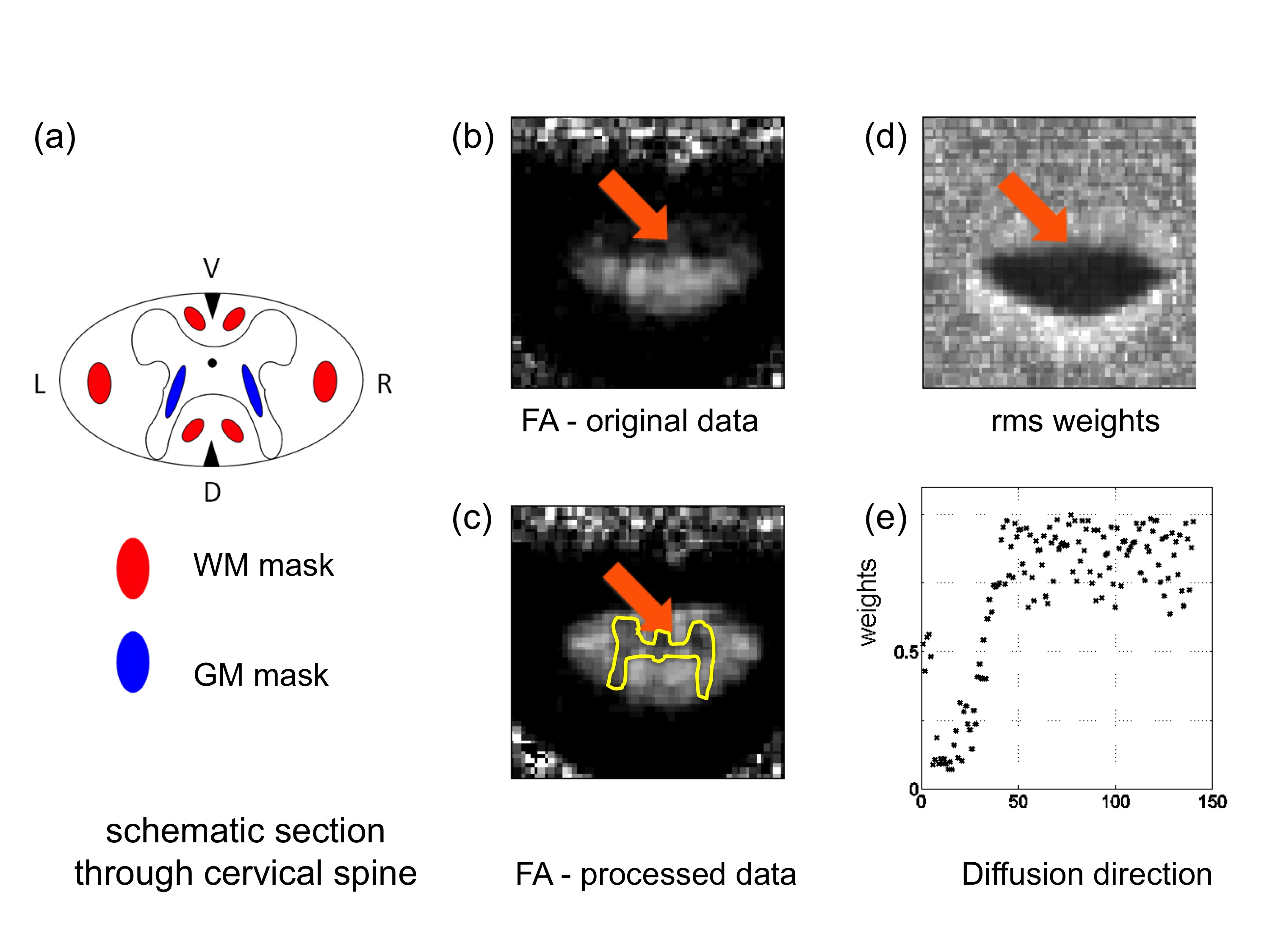
Fig. 2: (a) The spinal cord consists of white and gray matter and is a very small structure that is about 1cm in diameter. (b) Furthermore it is subject to motion and distortion artifacts that vary between different time-points during measurement and thus can bias spinal cord DTI (see arrow). (c) Robust Fitting can help to decrease this bias (arrow) by rejecting data points that were classified as outliers by the weights in Figure 1. (c) The root-mean-square (rms) of the weights clearly highlights the region in white matter where outliers lead to a bias in the spinal cord FA (arrow). (d) The weights for each single image tell us something about the confidence that we have in the corresponding data point.
Referencing
Mohammadi S, Freund P, Feiweier T, Curt A, Weiskopf N, The impact of post-processing on spinal cord diffusion tensor imaging, NeuroImage, Volume 70, 15 April 2013, Pages 377-385, doi: 10.1016/j.neuroimage.2012.12.058 .
Jan Malte Oeschger, Karsten Tabelow, Siawoosh Mohammadi, Axisymmetric diffusion kurtosis imaging with Rician bias correction: A simulation study bioRxiv 2022.03.15.484442;
(Please cite this paper when using this tool)
Updated