Wiki
Clone wikiACID - Artefact correction in diffusion MRI / T1wSusceptibility
Susceptibility distortion correction based on T1w
If no DTI-EPI with reversed phase-encoding gradients was acquired but a T1w of the same subject is available, susceptibility distortions can still be reduced using the board-tools that SPM provides. Here, we provide an example pipeline (Estimatefield_UnwarpDTI.mat) that can estimate a deformation field that reduces the susceptibility distortions in the b=0 image (see Figure 2-5). Furthermore, we present an example batch (ApplyDeformation2b0.mat) that could be used to apply the estimated field to the distorted b=0 image. The latter batch can also be applied to all other DTI images that were acquired within the same acquisition and the same DTI-protocol and thus have nearly the same susceptibility distortions as the b=0 image. Both batches are part of the ACID toolbox and can be found in the folder "/ExampleBatches/SPM12batches/".
Keep in mind that you need to replace the path of the T1w and b0 images by the path of your local T1w and b=0 images. You also need to add the "tissue-probability maps" (TPM) into the first Segmentation function - these are located in your local folder "spm12/tpm/".
Finally, we would like to emphasis that the T1w-based susceptibility distortion correction is not our recommended way, because it won’t perform as good as HySCO (see Figure 1). Furthermore, it should be mentioned that the parameters that were used in the "Estimatefield_UnwarpDTI.mat" batch are the default values provided by SPM. Probably, these parameters can be optimized to improve the performance of T1w-based susceptibility distortion correction method.
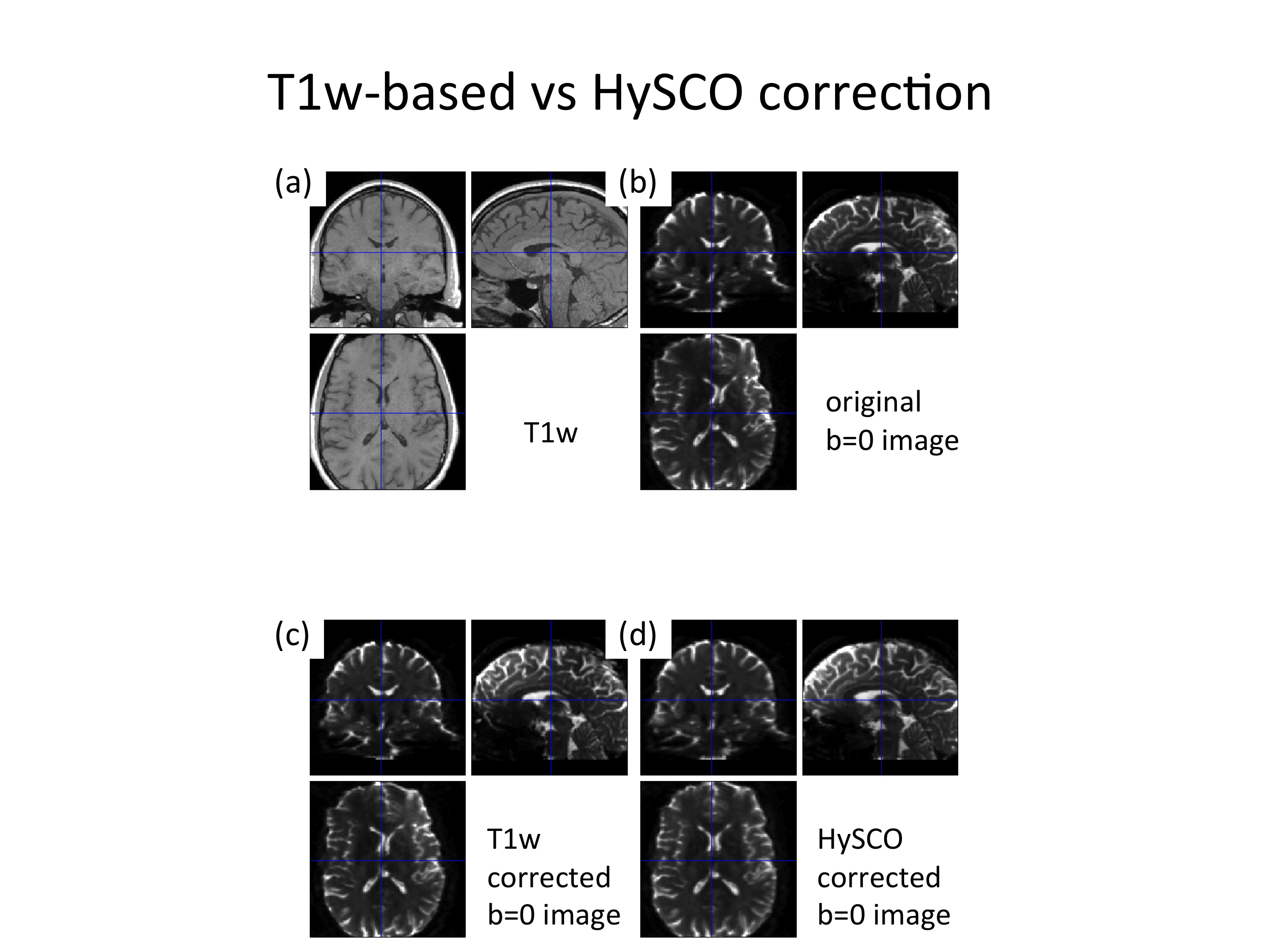
Figure 1: Example of susceptibility artifacts. As compared to the undistorted T1w image (a), the original b=0 image suffers from strong susceptibility distortions along the left-right direction (b). The T1w-image based susceptibility distortion correction can reduce these distortions to a certain degree (c) but does not perform as good as the HySCO-based correction (d).
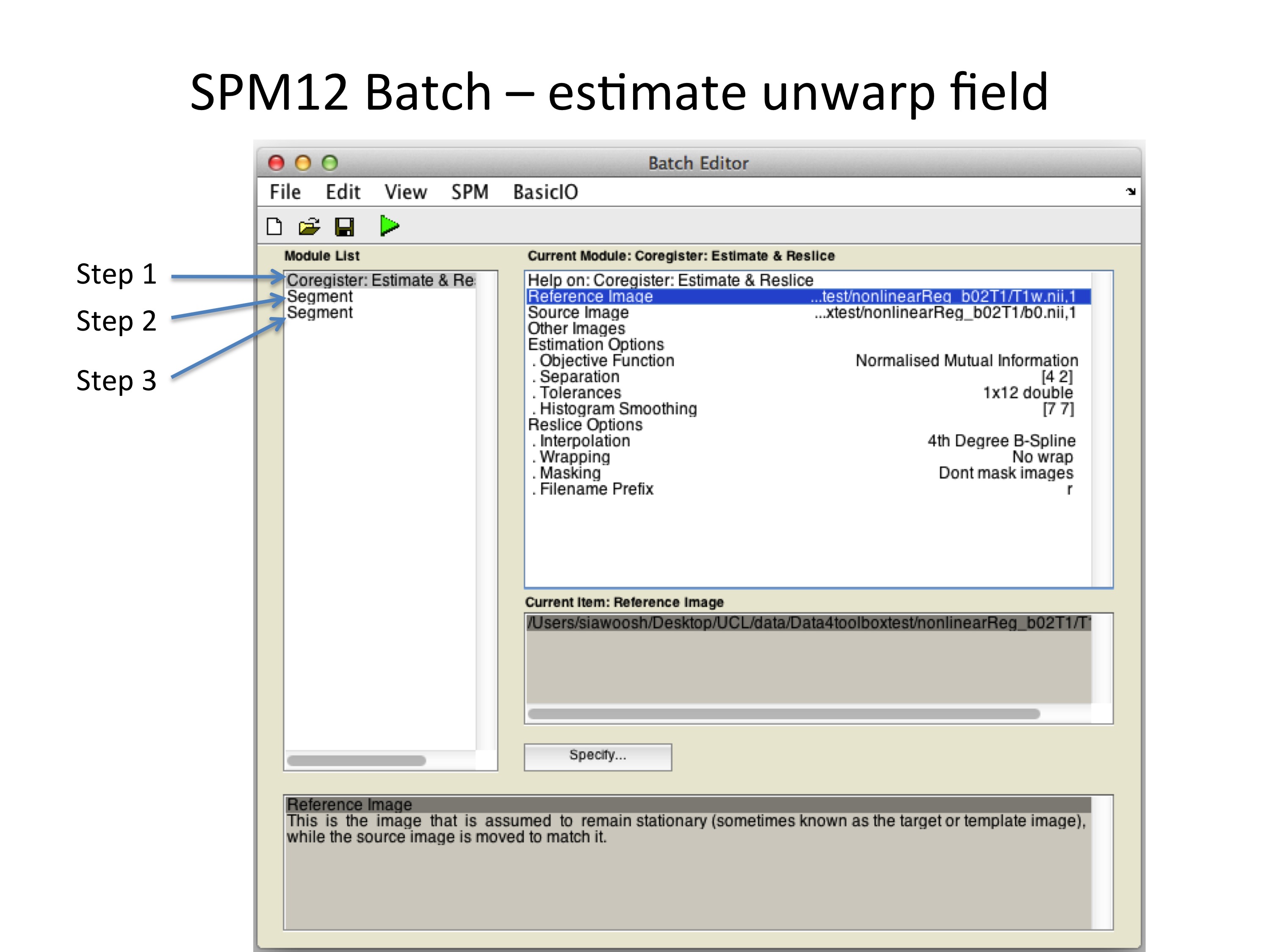
Figure 2: This batch uses SPM12 tools to register the b=0 image to the corresponding T1w image of an individual. It includes three steps, which will be described in more detail in the next figures.
Step (1): Coregister the b=0 image to the T1w image using “spm_coreg” to reduce potential inter-scan subject movement between acquisition of b=0 image and T1w image.
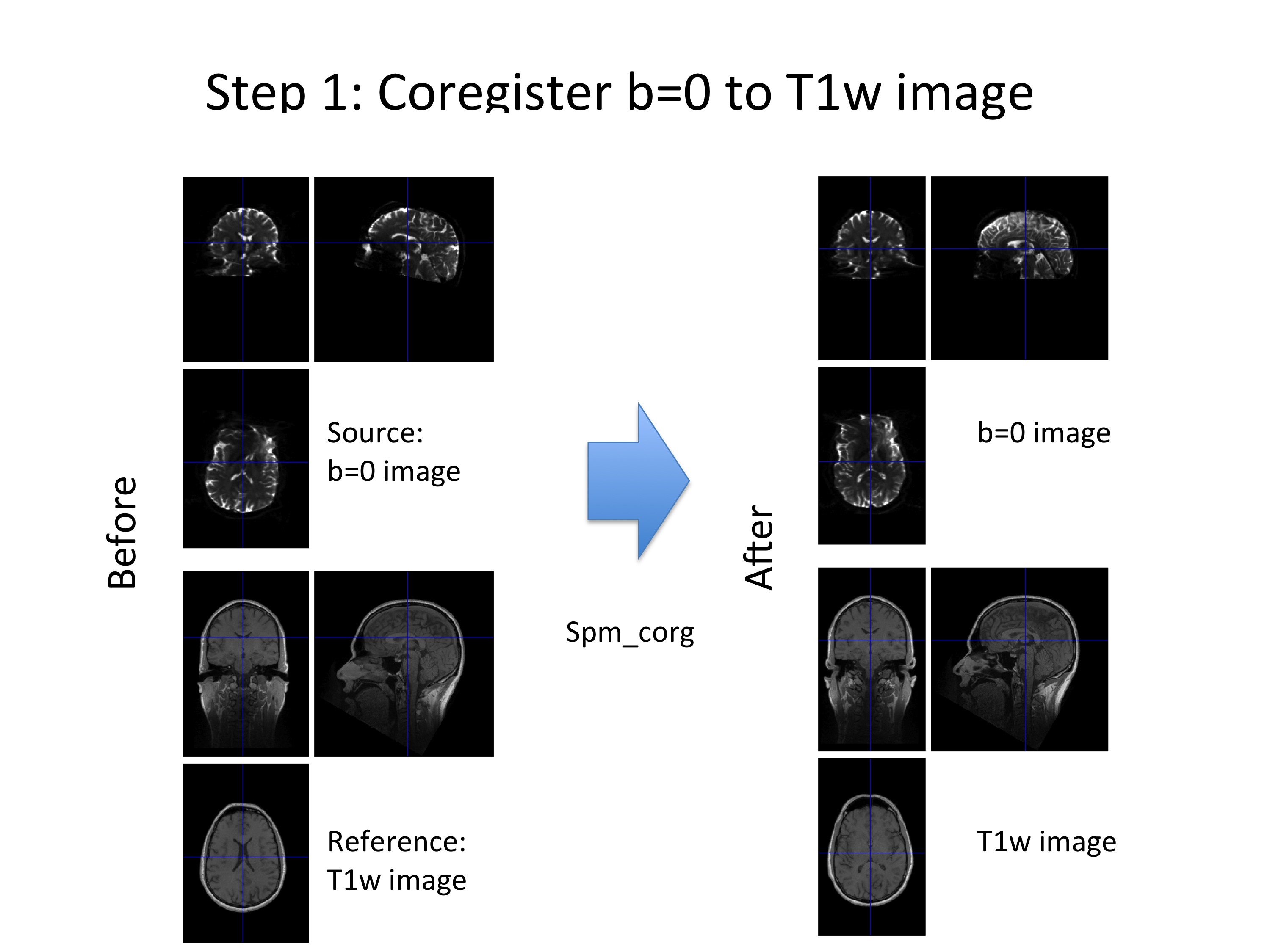
Figure 3: An example, where the subject moved between the acquisition of the b=0 and T1w images (left). This subject movement was corrected using spm_coreg (right).
Step (2): Segment T1w image using the default “spm_segment”.
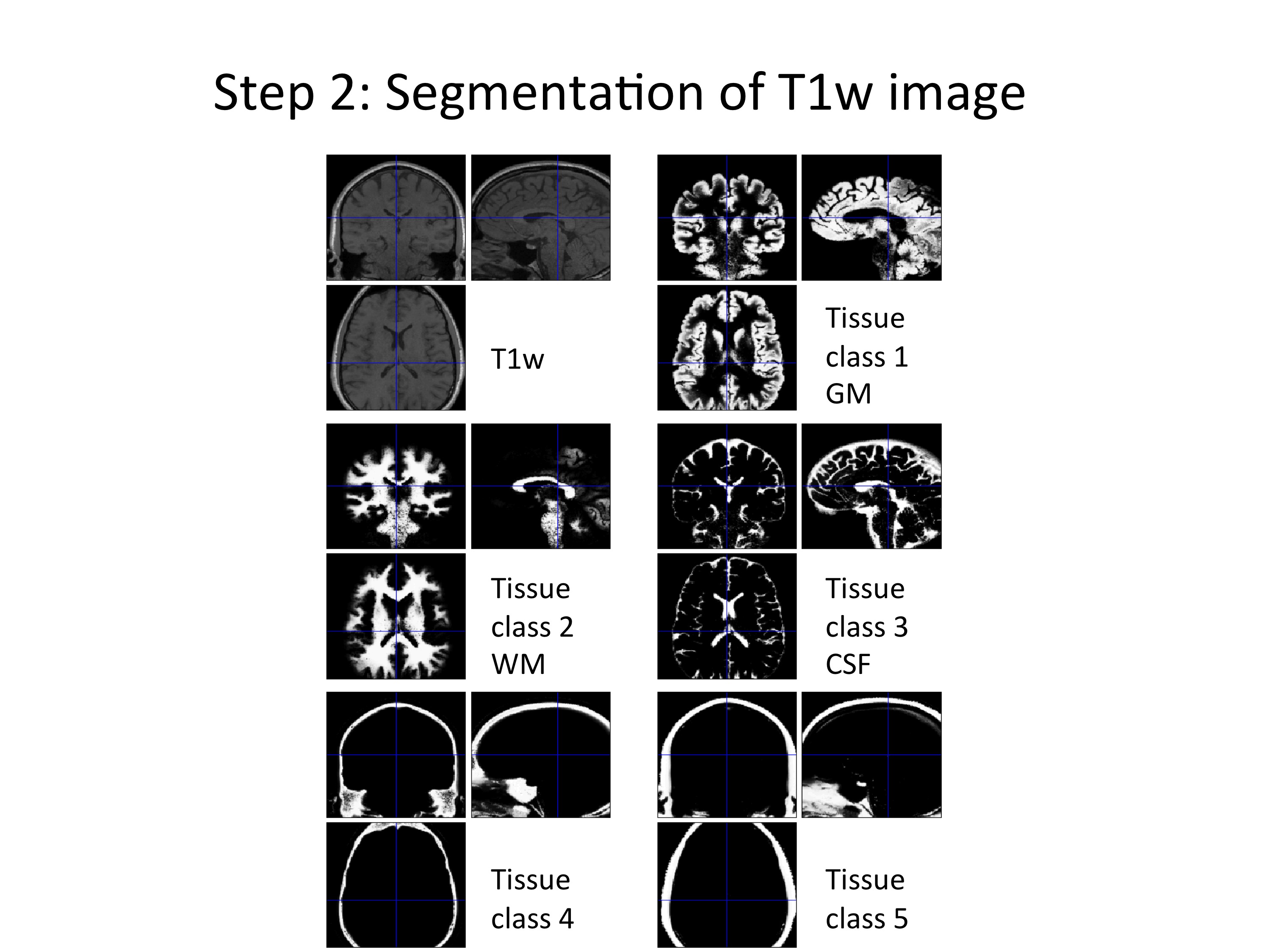
Figure 4: Example of segmenting the T1w image. Five different tissue classes were obtained from this step.
Step (3): Segment the b=0 image using the T1w probability maps obtained from step (2) as tissue probability maps and save forward and inverse deformation field.
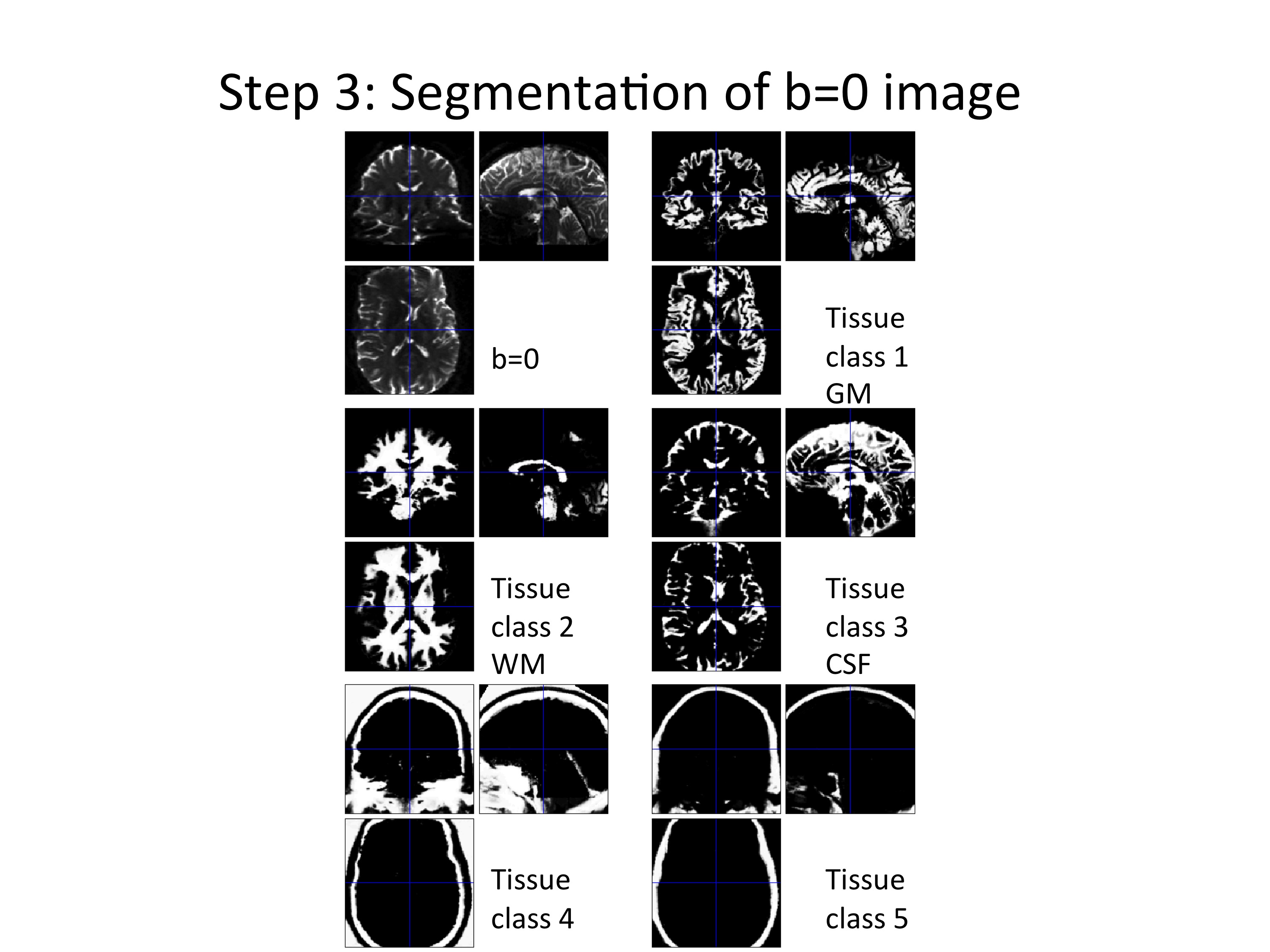
Figure 5: Example of segmenting the b=0 image. Five different tissue classes were obtained from this step.
Finally, the estimated deformation field can be used to unwarp the b=0 image via the deformation toolbox in SPM12 (Fig. 6).
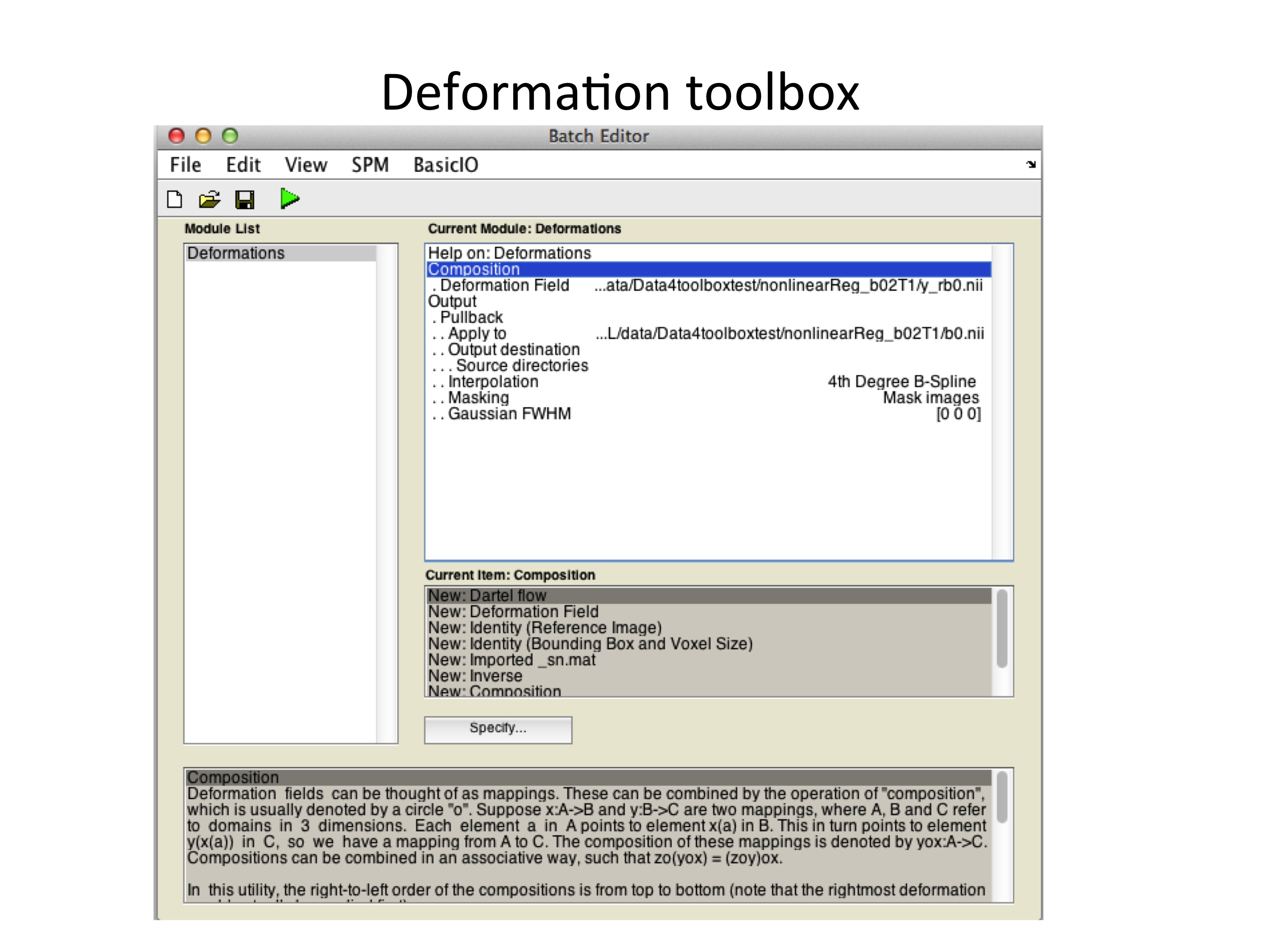
Figure 5: Example batch to unwarp the b=0 image using the deformation utility of SPM12.
Updated