Wiki
Clone wikienterobase-web / EnteroBase Backend Pipeline: refMasker
Top level links:
- Main top level page for all documentation
- EnteroBase Features
- Registering on EnteroBase and logging in
- Tutorials
- Using the API
- About the underlying pipelines and other internals
- How schemes in EnteroBase work
- FAQ
refMasker
Overview
refMasker identifies dispersed repeats, tandem repeats and CRISPR regions in assembly sequence (creating an annotation file in GFF format).
The refMasker pipeline is usually invoked as part of a workflow to create a SNP tree. An arbitrary assembly is chosen as a reference in that workflow and refMasker is used to identify sequence that is problematic for later steps in the workflow.
refMasker is currently in version 1.0.
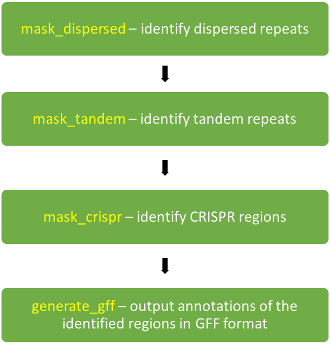
Dispersed repeats
Dispersed repeats are identified in a BLASTN (version 2.2.31) search (using the command line executable), using a database created with makeblastdb from the assembly sequences, in order to identify where there are sequence matches (with trivial matches due to sequences matching themselves at the same location screened out).
Tandem repeats
Tandem repeats are identified using trf (version 4.07 beta).
CRISPR regions
CRISPR regions are identified using pilercr (version 1.06).
Updated